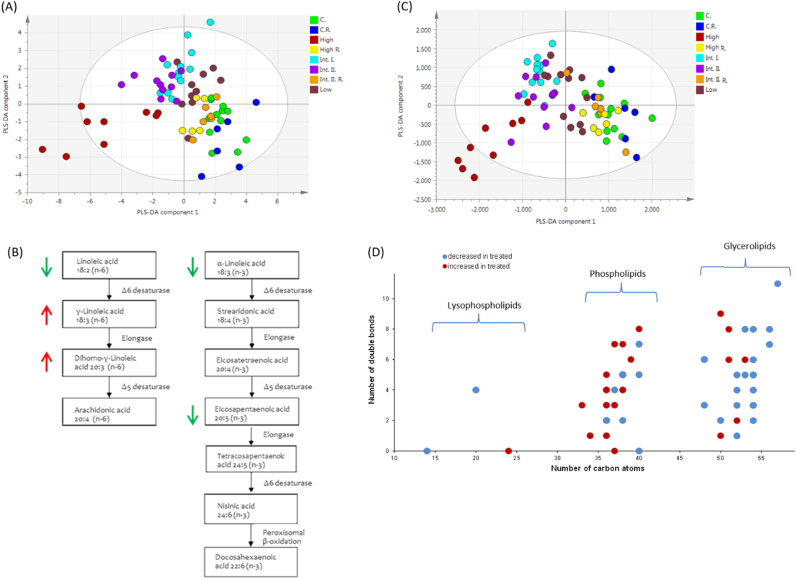
Fig. 2.
Overview of gas chromatography–mass spectrometry (GC–MS) and liquid chromatography–mass spectrometry (LC–MS) data from the lipid extracts of rat liver tissue samples treated with the PPAR-pan agonist GW625019. (A) Partial least squares-discriminant analysis (PLS-DA) scores plot shows total fatty acid data from lipid extracts of liver tissue samples with clear clustering associated with dose of the agonist. (B) Metabolic changes caused by PPAR-pan agonist GW625019 are increased γ-linoleic acid (GLA) and dihomo-γ-linoleic acid (DGLA). Linoleic acid (LA), α-linoleic acid (ALA) and eicosapentaenoic acids (EPA) concentration were decreased. Arrows show the direction of changes in lipid concentrations relative to control samples in red (increased) and green (decreased) respectively. (C) PLS-DA scores plot of LC–MS intact lipid data (D) and an overview of the chain length and the saturation of changing lipids. Red circles represent the compounds which have increased, whereas blue circles show decreased lipid species. Treatment abbreviations: C., control; Int. I., intermediate 1; Int. II., intermediate 2; R., recovery.