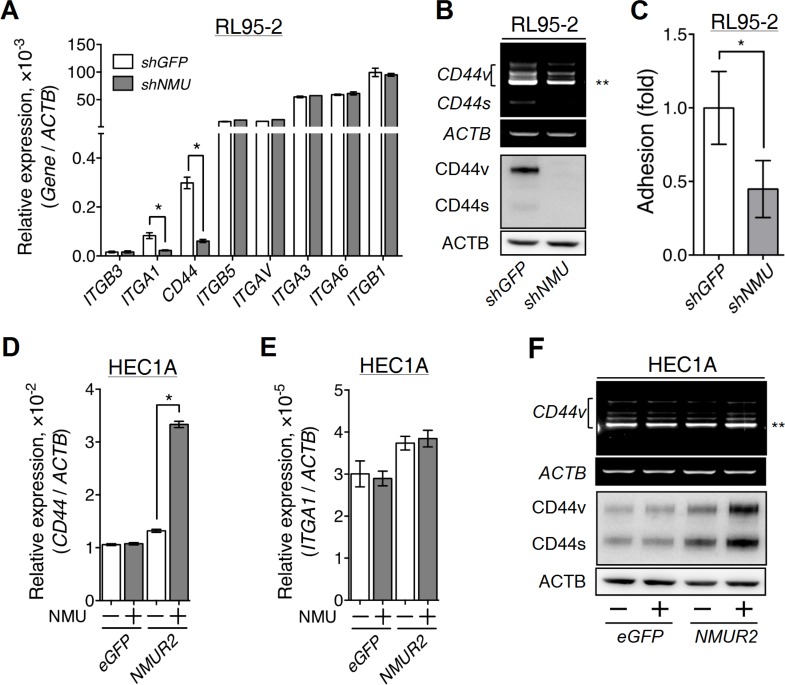
Figure 5. Effects of NMU signaling on the expression of adhesion molecules in the grade II endometrial cancer cell lines.
(A) The transcript levels of selected adhesion molecules were quantified in RL95-2 cells without or with NMU knockdown. (B) A primer pair across non-variable exons 5 and 7 of CD44 (forward, CACCCCATCCCAGACGAA; reverse, CCACCTTCTTGACTCCCATGT) was used to unveil the transcript profiles of CD44 splicing variants. The transcript profiles (upper panel) and protein profiles (lower panel) of standard CD44 (CD44s) and its splicing variants (CD44v) were revealed in RL95-2 cells without or with NMU knockdown. (C) RL95-2 cells without or with NMU knockdown were seeded on the hyaluronan-coated plate to compare their matrix adhesion ability. In addition, the transcript levels of (D) CD44 and (E) ITGA1, and (F) the splicing profiles (upper panel) and protein profiles (lower panel) of CD44 were also compared between eGFP-overexpressing and NMUR2-overexpressing HEC1A cells without or with 100 nM NMU treatment. The transcript and protein levels of ACTB served as controls. Quantification data are shown as the mean ± SD. *p < 0.05. **CD44v8-10, the major splicing variant of CD44 gene in RL95-2 and HEC1A cells.