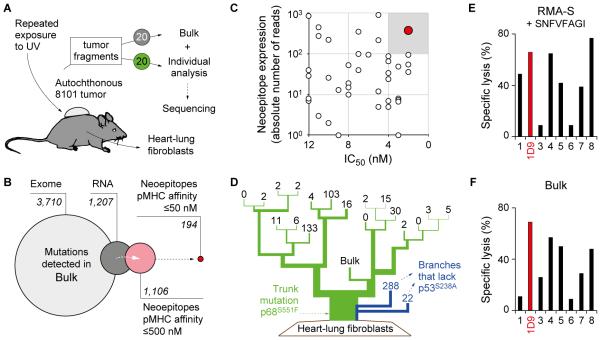
Fig. 1. The neoepitope mp68, predicted by ‘reverse immunology’, is found throughout the genetically diverse tumor 8101 and is recognized by high-avidity T cell clones.
A, UV-irradiation caused the development of an autochthonous tumor (8101). The tumor was excised and 20 individual tumor fragments were adapted to culture (Bulk) or analyzed separately using whole exome sequencing. Heart-lung fibroblasts (HLF) were generated as autologous tissue control. B, Identification of suitable neoepitopes as therapeutic targets by ‘reverse immunology’. Venn diagram (from left to right): Number of mutations detected in Bulk after whole exome and RNA sequencing. Number of neoepitopes (8-, 9-, or 10-mer peptides) found to be expressed and predicted to bind to H-2Kb or -Db with affinities of ≤500 nM or ≤50 nM (NetMHC 3.4). C, Binding affinity to MHC-I (IC50) and RNA expression level of neoepitopes identifies mp68 (SNFVFAGI, red) as highly expressed antigen with highest MHC affinity. D, Phylogenetic representation of somatic mutational frequency in the 8101 tumor identifies mp68 as trunk mutation. Green represents the trunk mutation p68S551F that was found in all fragments. Branches shown in blue lack the mutation p53S238A. Numbers on the top of each branching indicate unique mutations in 20 individual fragments and the Bulk tumor cell culture of 8101. E, F, The T cell clone 1D9 efficiently lyses Bulk tumor cells and is specific for the mp68 neoepitope. Specific lysis of (E) RMA-S cells loaded with 7.8 pM SNFVFAGI peptide or (F) Bulk tumor cells by mp68-specific T cell clones. The T cell clone 1D9 is highlighted in red and was used for subsequent TCR isolation.