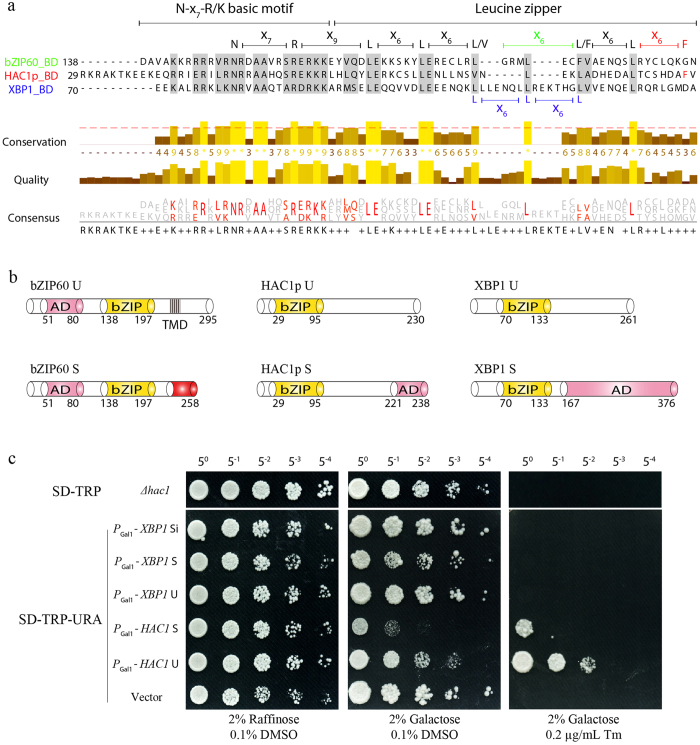
Figure 1. XBP1 is not Functionally Complementary with HAC1p in Yeast.
(a) The conservation and divergence of the bZIP domain between HAC1p, bZIP60, and XBP1. The residues with a conservation threshold of >8 (red dashed line) were shaded in grey. The conservation quality and consensus sequence were given below with a histogram. A schematic of the bZIP domain consensus was shown above by extremely conserved residues and distance. Note that the N-x7-R/K basic motif is conserved in HAC1p, bZIP60 and XBP1. The zipper motif consists of five heptad Leucine-repeats in XBP1, whereas four in HAC1p and bZIP60 with and without interruption, respectively. (b) Although the AD locus in bZIP60 is different from that in HAC1p, their AD domains share a high identify in amino acid sequence26. However, the ADs of XBP1 S and HAC1p S are greatly different in length and in amino acid sequence, even though they both result from the unconventional splicing and have the same locus. (c) Functional testing of XBP1 in CRY1 Δhac1::TRP strains. The untransformed or transformed yeast cells with empty CEN-ARS plasmid or plasmid harboring HAC1 S, HAC1 U, XBP1 U, XBP1 S and XBP1 Si were normalized to an OD600 = 0.3 after 8 h of growth in raffinose-containing media, and 5-fold serial dilutions were spotted on raffinose and galactose-containing plates supplemented with 0.1% DMSO (control) or 0.2 μg/mL Tm. Plates were photographed after 2–3 days at 30 °C.