Fig. 4.
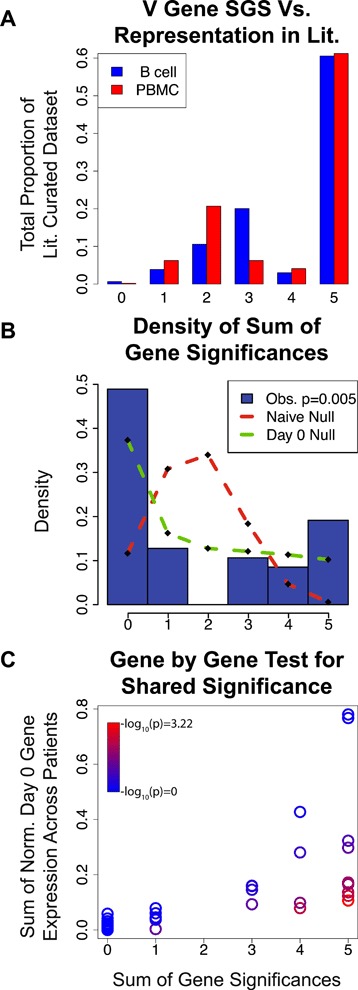
Identifying a convergent V gene usage signal across patients. The x-axis for all plots is the sum of gene significances (SGS) statistic, which is the number of patients for which a given Ab gene was found to be significant. a Comparing our results for IGHV to the literature. For each SGS bin, this shows the proportion of the Abs in the literature-curated data that have V genes belonging to this bin. Approximately 60 % of the influenza binding Abs in the literature-curated dataset were composed of V genes that had an SGS value of 5. b Comparing observed SGS to the null distribution. Blue bars are a histogram showing the observed proportion of IGKV genes from the PBMC data belonging to each SGS bin. Red dashed line shows the “naïve” null distribution of SGS if each patient were independent from one another (see “Methods”). The green dashed line shows the null distribution of SGS if the baseline similarity in gene expression at day 0 is taken into account. The p value in the legend shows the result of using a multinomial G test to compare the observed SGS distribution to that of the day 0 null. c Comparing the SGS value for each IGKV gene from the PBMC data to that of their respective null expectations. Color indicates the probability of the observed SGS under the null model (p value, see “Methods” for explanation of null model)
