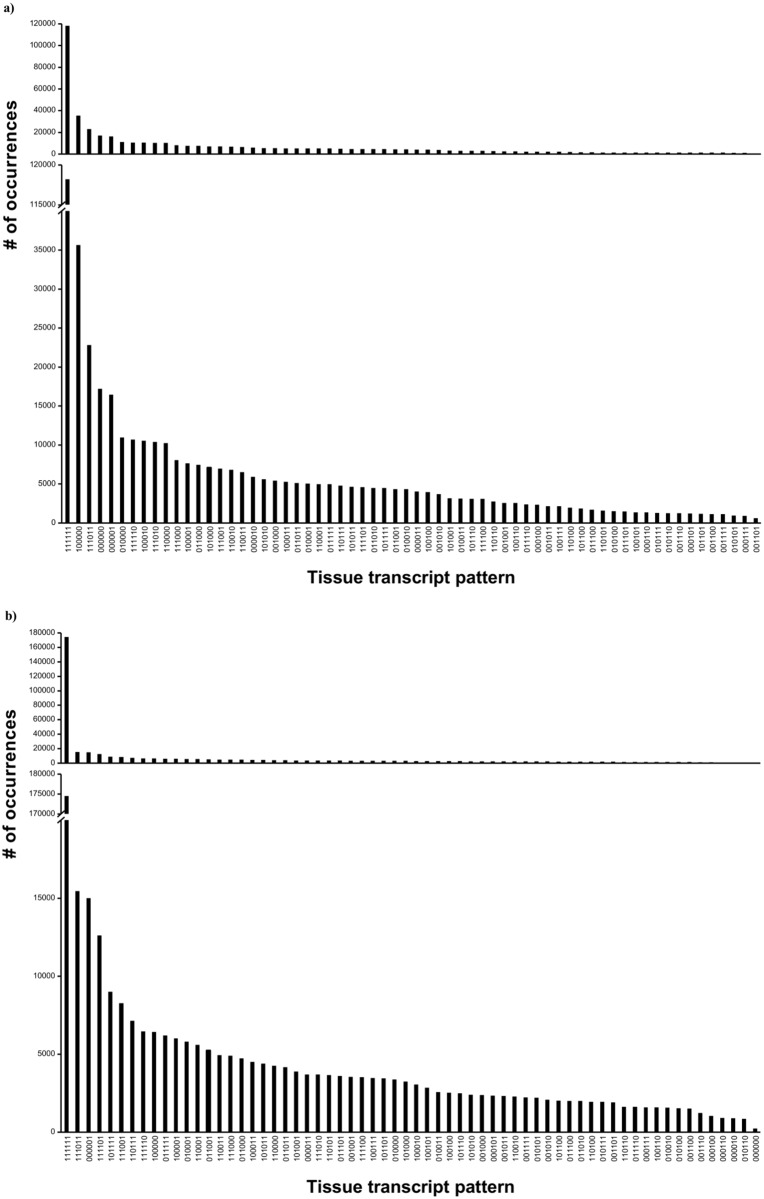
Fig 1. Transcript patterns across six different S. glomerata tissues.
A) non-normalised and non-strand specific tissue reads were mapped to the combined reference transcriptome and b) strand-specific and normalised tissue reads were mapped to the strand-specific reference transcriptome, using CLC Workbench version 7.5. Graph shows the number of transcripts that are common across individual tissue patterns, with 1 standing for “transcript present in tissue” and 0 for “transcript not present in tissue”. The order in which the tissues are listed is as follows: haemolymph (closest to the x-axis label), gill, mantle, muscle, gonad and digestive. The respective top graphs show the full-scale graph, with the respective lower graphs emphasising the fine scale features of the same graph by adding a graph break.