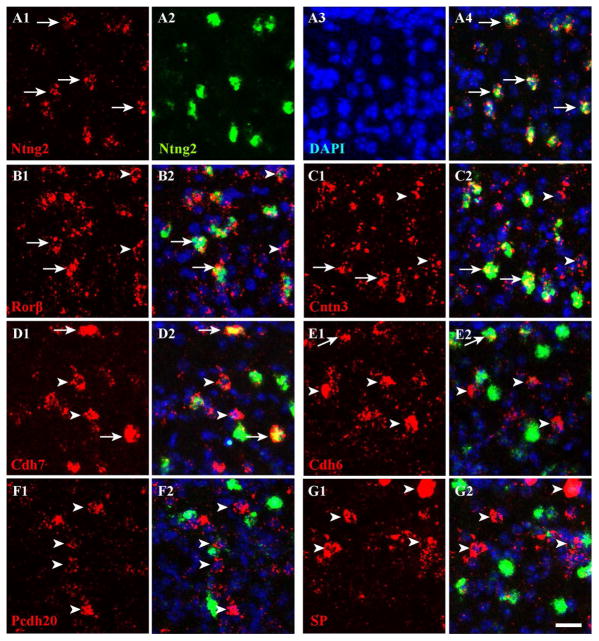
Figure 9. Ntng2 marks a subpopulation of Rorβ+ neurons in sSC.
A–G. The dFISH was conducted in sSC at ~P10 using Ntng2 as a second probe (labeled with fluorescein, green) and counterstained with DAPI (blue). The first probes (labeled with digoxigenin) are shown in red. All images in this figure were acquired in layer 1. A. Two in situ probes to Ntng2 (A1 and A2) highly overlapped (merged, A4; DAPI alone, A3). Here and in subsequent panels, arrows indicate examples of cells with overlapping expression, and arrowheads indicate expression by the first probe only. Arrows are missing in F and G because overlapping expression between the two genes is rare (<8%). B–D. Approximately half of Rorβ+ (B1), Cntn3+ (C1) and Cdh7+ (D1) cells were Ntng2+ (merged; B2, C2, D2). E–G. Very few cells expressing Cdh6 (E1), Pcdh20 (F1) and SP (G1) were Ntng2+ (merged; E2, F2, G2). Scale bar: 20 μm.