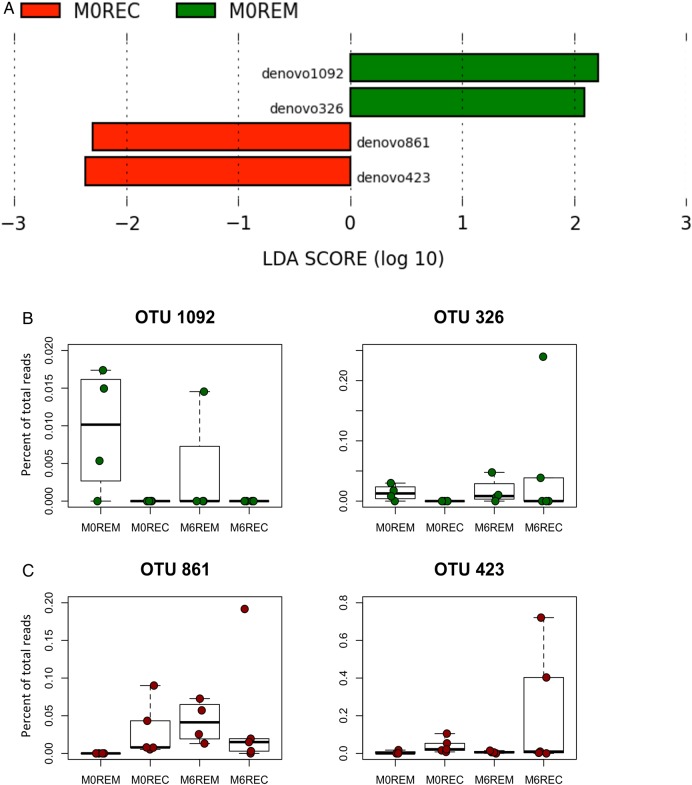
Figure 2.
Early bacterial biomarkers of the clinical outcome after ileocolonic resection (ICR). Faecal microbiota from 10 patients was analysed at M0 and 6 months after ICR. (A) Specific bacterial operational taxonomic units (OTUs) that may predict at M0 the clinical outcome following ICR were deciphered applying the LEfSe algorithm.47 The LEfSe algorithm uses the non-parametric factorial Kruskal–Wallis sum-rank test to detect features with significant differential abundance with respect to the clinical outcome; biological significance is subsequently investigated using a set of pairwise tests using the (unpaired) Wilcoxon rank-sum test. As a last step, LEfSe uses linear discriminant analysis to estimate the effect size of each differentially abundant feature (ie, bacterial OTUs). (B) Boxplot of the distribution (per cent of total reads) of the four OTUs highlighted in the LEfSe analyses in the different patients’ groups. M0 bacterial biomarkers associated with remission at M6 are in green. M0 bacterial biomarkers of future recurrence in patients undergoing ICR are in red. M0, samples at time of surgery; REC, recurrence; REM, remission; LDA, linear discriminant analysis.