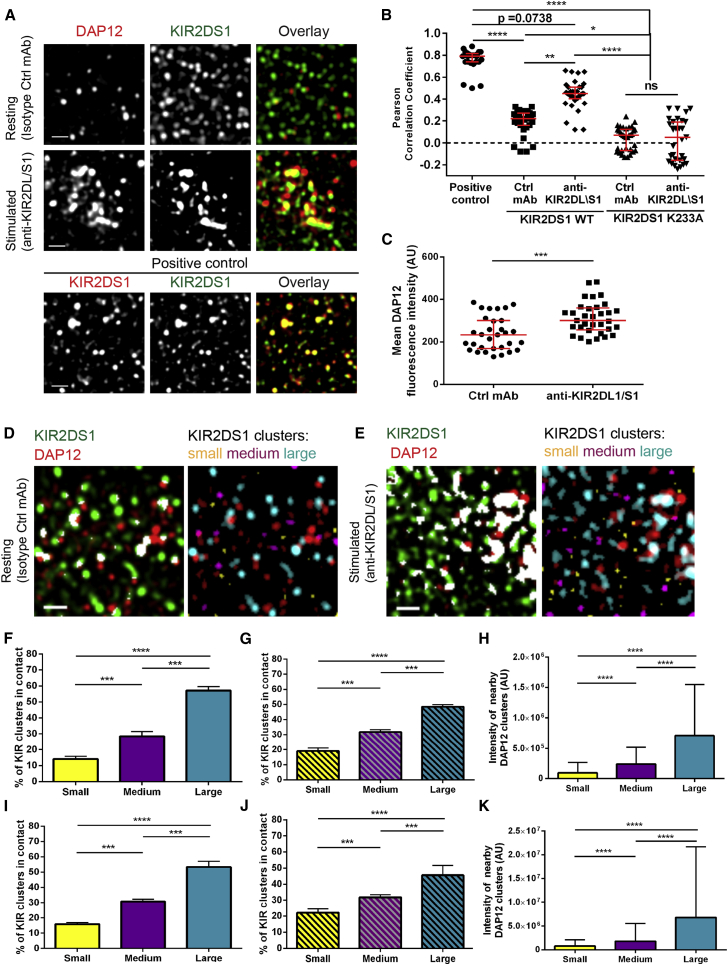
Figure 5.
Interaction between Nanoclusters of KIR2DS1 and DAP12
(A) NKL/KIR2DS1-HA cells were incubated on isotype control-coated (top row) or anti-KIR2DL/S1 mAb-coated (middle row) slides, stained for KIR2DS1 (with an anti-HA mAb, AF488) and DAP12 (AF568), and imaged with STED. For a positive control (bottom row), NKL/KIR2DS1-HA was stained with anti-KIR2DL/S1 mAb (green; Atto488) and anti-HA mAb (red; AF568). Representative 3 × 3 μm regions from STED images of NKL/KIR2DS1-HA cells with channels overlaid and separated (overlapping green and red pixels are marked yellow). Scale bars represent 500 nm.
(B) Pearson correlation coefficients for KIR2DS1 or KIR2DS1K233A and DAP12 in cells on isotype control-coated and anti-KIR2DL/S1 mAb-coated slides, compared to positive control.
(C) Mean DAP12-derived fluorescence intensity in NKL/KIR2DS1-HA cells on isotype control- and anti-KIR2DL/S1 mAb-coated slides. In (B) and (C), each symbol represents one cell. Horizontal bars and errors represent the medians and interquartile range.
(D–K) Quantification of interaction between differently sized KIR2DS1 clusters and DAP12 clusters in NKL/KIR2DS1-HA cells incubated on isotype control-coated (D, F–H) and anti-KIR2DL/S1 mAb-coated slides (E, I–K). In (D) and (E), 3 × 3 μm regions of cell membrane with KIR2DS1 (green) and DAP12 (red) are shown with overlapping pixels marked in white (left) or with KIR2DS1 clusters color-coded according to their size (right). Each size bin corresponds to one-third of all KIR2DS1 clusters. (F and I) The fractions of all KIR2DS1 clusters associated with DAP12 that fall into each size group. (G and J) The fractions of all KIR2DS1 clusters associated with DAP12 that fall into each size group in simulated data where the positions of KIR2DS1 clusters were maintained and positions of DAP12 clusters were randomized within the boundaries of cell area. (H and K) Average intensity of DAP12 clusters found nearby KIR2DS1 clusters from specified size bins. (F–K) Bars and errors represent the medians and interquartile range. Data are from 25–34 cells from two independent experiments. AU, arbitrary units; ns, non-significant. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Kruskal-Wallis test by ranks with Dunn’s post-test (B), Mann-Whitney test (C), row-matched Friedman test with Dunn’s post-test (F–K).