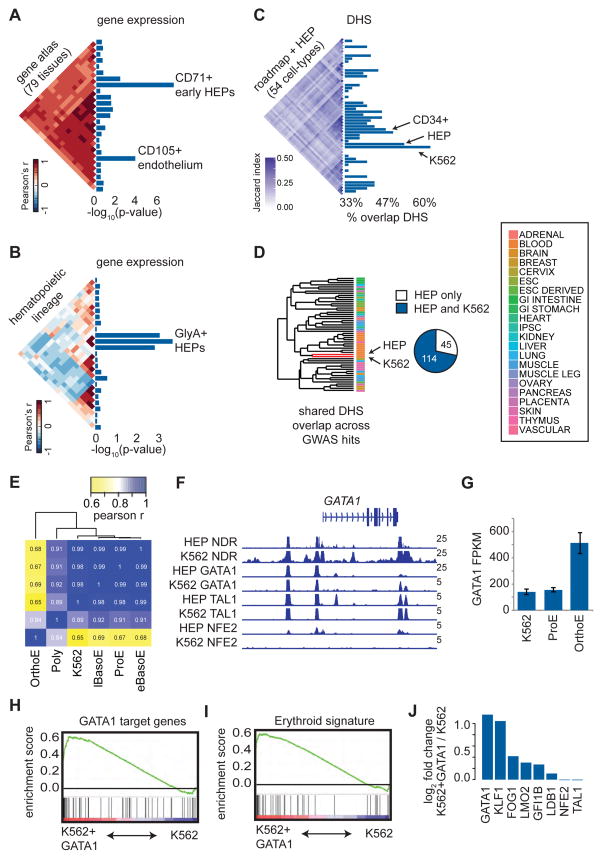
Figure 2. Red blood cell trait associated variants are enriched for erythroid regulatory regions.
(A–B) Cell type enrichment for specifically expressed genes proximal to GWAS hits. Triangular heatmaps of tissue similarity are shown for the Pearson correlations of each tested tissue set and enrichment scores are represented as −log10 p-values. Only cell types reaching a minimum enrichment are plotted. (C) Similarity of DHS peaks across 54 cell types is shown as a triangular heatmap of the Jaccard statistic. Bar plots show the percentage of GWAS hits such that at least one variant in high LD with each tag SNP overlaps with open chromatin in each of the 54 cell types. (D) Regions of open chromatin in HEPs and K562 cells are highly overlapping based upon hierarchical clustering of the 54 cell types in (C). Nearly 75% of the variants in high LD with GWAS hits that fall within open chromatin in HEPs are also in open chromatin in K562 cells. (E) Based upon RNA-seq, K562 gene expression is highly correlated (Pearson r) with that of early HEPs (proerythroblasts, ProEs, and basophilic erythroblasts, BasoEs), while more mature HEPs (polychromatic erythroblasts, PolyEs, and orthochromatic erythroblasts, OrthoEs) show distinct expression profiles. (F) Occupancy by GATA1, TAL1, NFE2, and nucleosome-depleted regions (NDRs) across the GATA1 locus is similar for both HEPs and K562 cells. (G) GATA1 gene expression is similar between early HEPs and K562 cells but increases substantially in mature HEPs. (H and I) Gene Set Enrichment Analyses (GSEA) indicate that K562 cells overexpressing GATA1 exhibit an increased expression of GATA1 target genes, resulting in an overall more mature erythroid gene expression signature. The enrichment score is plotted from GSEA. (J) Upon overexpression of GATA1, multiple erythroid TFs and GATA1 co-factors are upregulated in K562 cells.