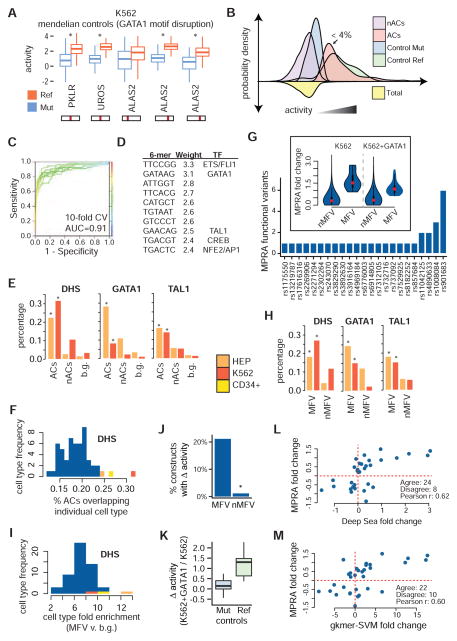
Figure 3. Identifying erythroid regulatory elements and MPRA functional variants.
(A) Activity boxplots of the 5 unique positive control variants. Constructs with intact GATA1 binding sites (Ref) show increased activity when compared with broken binding sites (Mut). (B) Kernel densities for positive controls as well as for ACs and nACs containing GWAS variants. ACs represent 4% (555/15612) of the MPRA library. (C) Presence or absence of specific 6-mers can effectively be used to discriminate between ACs and nACs using a support vector machine model. (D) The 6-mers that are most strongly weighted towards ACs are similar to ETS/FLI1, GATA1, TAL1, CREB, and NFE2/AP1 motifs. (E) ACs are enriched for erythroid DHS as well as for occupancy sites of the erythroid TFs, GATA1 and TAL1, when compared to low activity constructs (nACs) and ~10,000 background sentinel GWAS hits. (F) Overlap of ACs with sites of open chromatin across multiple cell types. (G) 32 MPRA functional variants (MFVs) representing 23 GWAS hits (median 1 / GWAS hit) were identified based upon differential activity between the major and minor alleles. (insert) Absolute fold change sizes comparing construct pairs meeting the 1% FDR cutoff for MFVs vs. all other constructs. (H) Similar to (E), except for MFVs. (I) Similar to (F), except for MFVs and the enrichment is computed for MFVs compared to ~10,000 background GWAS hits. (J) The group of MPRA functional variant (MFV) constructs is significantly enriched for constructs with dosage-dependent GATA1 activity. (K) Ref, but not Mut, GATA1 binding sites show increased activity upon GATA1 overexpression. (L) Correlations between MRPA and DeepSea (trained on K562 DNase I hypersensitivity) fold change is shown for all MFVs. MPRA fold change was calculated as the mean across all sliding windows (K562+GATA1 fold change shown). (M) Similar to (L), except for the gkmer-SVM algorithm.