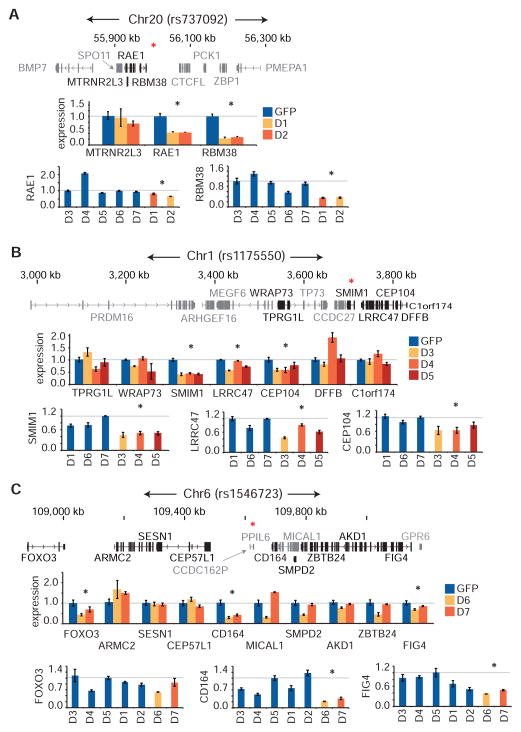
Figure 5. Altered gene expression in clonal isogenic deletions of rs737092, rs1175550, and rs1546723.
(A–C) Small clonal deletions were made in K562s across rs737092 (clones D1, D2), rs1175550 (clones D3, D4, D5), and rs1546723 (clones D6, D7) using CRISPR-Cas9 genome editing. Control clones were generated by transfecting Cas9 and pLKO.1 GFP constructs. Quantitative RT-PCR analysis of genes within a ~1 megabase wingspan of rs737092 (A), rs1175550 (B) and rs1546723 (C) and expressed over a minimum threshold (FPKM > 2, genes not meeting threshold are in grey) in HEPs was performed. A Bonferroni correction was applied at each locus. A red asterisk (*) indicates the position of the MFV.