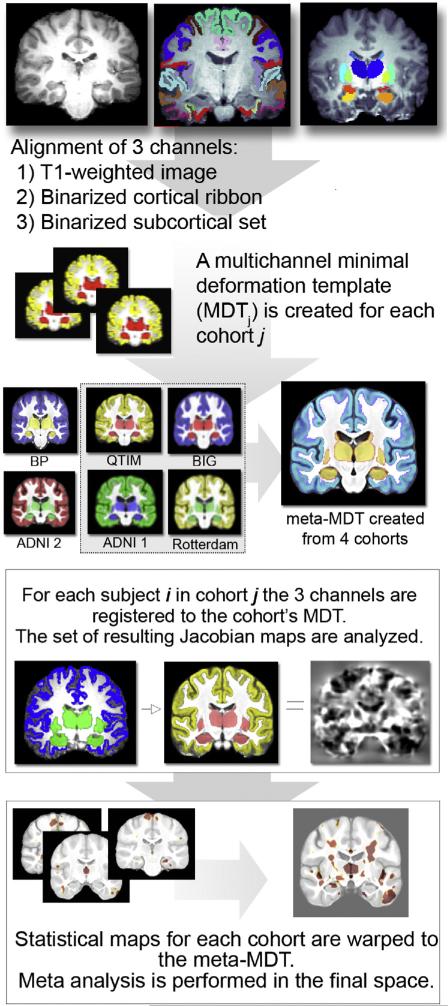
Fig. 5. Meta-Analyzing Statistical Brain Maps.
As in other fields of brain mapping, voxel-based statistical analyses can map statistical associations between predictors and brain signals. To meta-analyze maps of statistical associations across sites, Jahanshad et al. (2015a,b,c) proposed a method whereby each site aligns data to their own brain template (mean deformation template, or MDT). Statistics from each site are meta-analyzed at each voxel, after a second round of registration to an overall mean template (computed here from 4 cohorts representing different parts of the lifespan). Analyses proceed in parallel, using computational resources across all sites; analyses are updated when a new site joins. This approach applies equally to voxel-based maps of function, and the ENIGMA-Shape working group has modified it to work with surface-based coordinates (Gutman et al., 2015a,b,c). If structural labels are used to drive the multi-channel registration (top panels), in conjunction with an approach such as tensor-based morphometry, the resulting local volumetric measures should closely mirror volumetric findings for specific regions of interest. As such, some results of brain-wide genome-wide searches can be checked by consulting genome-wide association results for specific regions of interest (Hibar et al., 2015a,b; Adams and the CHARGE and ENIGMA2 Consortia, submitted for publication).