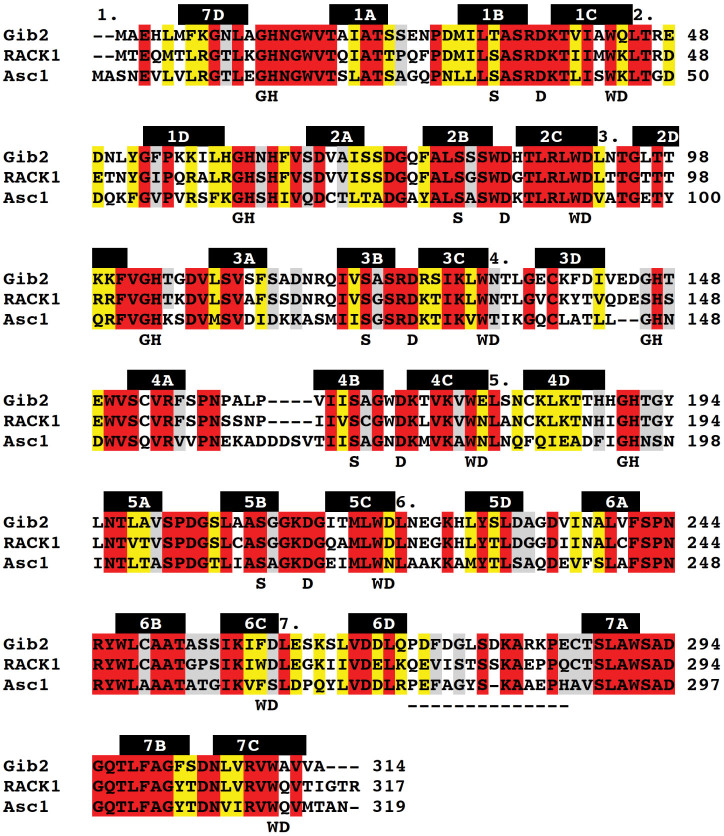
Figure 1. Sequence alignment of C. neoformans Gib2 (UniProt accession number A0AUJ0), H. sapiens RACK1 (P63244), and S. cerevisiae Asc1 (P38011).
Multiple sequence alignment was performed using ClustalW2 (v2.1). All fully conserved residues are highlighted in red. Conserved residues with high (scoring > 0.5 in the Gonnet PAM 250 matrix) and moderate (scoring ≤ 0.5) similar properties are highlighted in yellow and grey, respectively. The WD motifs are numbered above the sequences, and the positions of the conserved WD and GH repeats, as well as structurally conserved S and D residues in WD proteins are shown below the sequences. The locations of β-sheets forming the propeller blades in Gib2 are indicated above the sequence as black bars, and the extended loop residues are highlighted in the dashed line below the sequence.