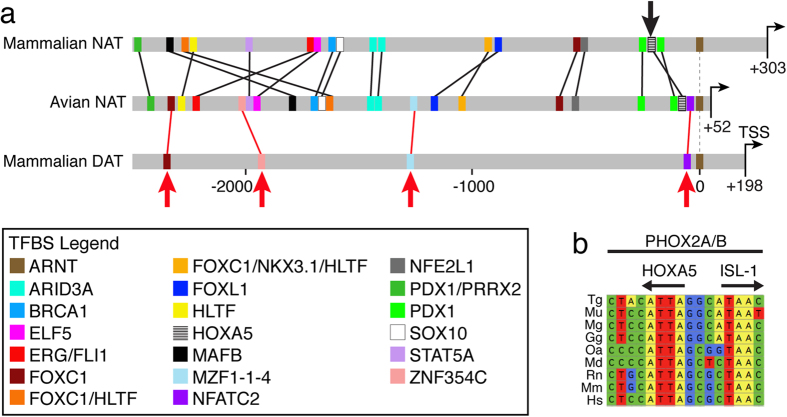
Figure 4. Comparative analysis of NAT and DAT cis-regulatory promoter elements.
(a) Transcription factor binding sites (TFBSs; colored vertical rectangles) that are conserved in avian NAT (middle line) compared with mammalian NAT (top line) or DAT (bottom line). Shown are ~2,500 kb upstream of the putative transcription start sites (TSSs, thin horizontal arrows), and only the TFBSs that are conserved in all organisms examined within each lineage and that are shared between avian NAT and mammalian NAT (black connecting lines), or avian NAT and mammalian DAT (red connecting lines and red arrows; see Methods for details). Promoters are aligned with respect to the position of the zebra finch and mouse ARNT (indicated by thin dashed line), a conserved site immediately upstream of the putative TSSs in all NAT and DAT promoters. The black vertical arrow indicates the position of a homeodomain-binding motif known to confer cell-type specific expression to human NAT in noradrenergic neurons59 and found to be conserved and shared between avian and mammalian NAT, and not mammalian DAT promoters. Color key to all TFBSs is given by legend on bottom left. (b) Genomic alignments of a short stretch of the NAT promoter derived from a set of representative species indicates that the core human NAT proximal homeodomain motif that contains binding sites for HOXA5, ISL1 and PHOX2A/B, is conserved across vertebrate phylogeny. Species name abbreviations: Tg, Taeniopygia guttata, Mu; Melopsittacus undulatus, Mg, Meleagris gallopavo; Gg, Gallus gallus; Oa; Ornithorhynchus anatinus; Md, Monodelphis domestica; Rn, Rattus norvegicus; Mm, Mus musculus; Hs, Homo sapiens.