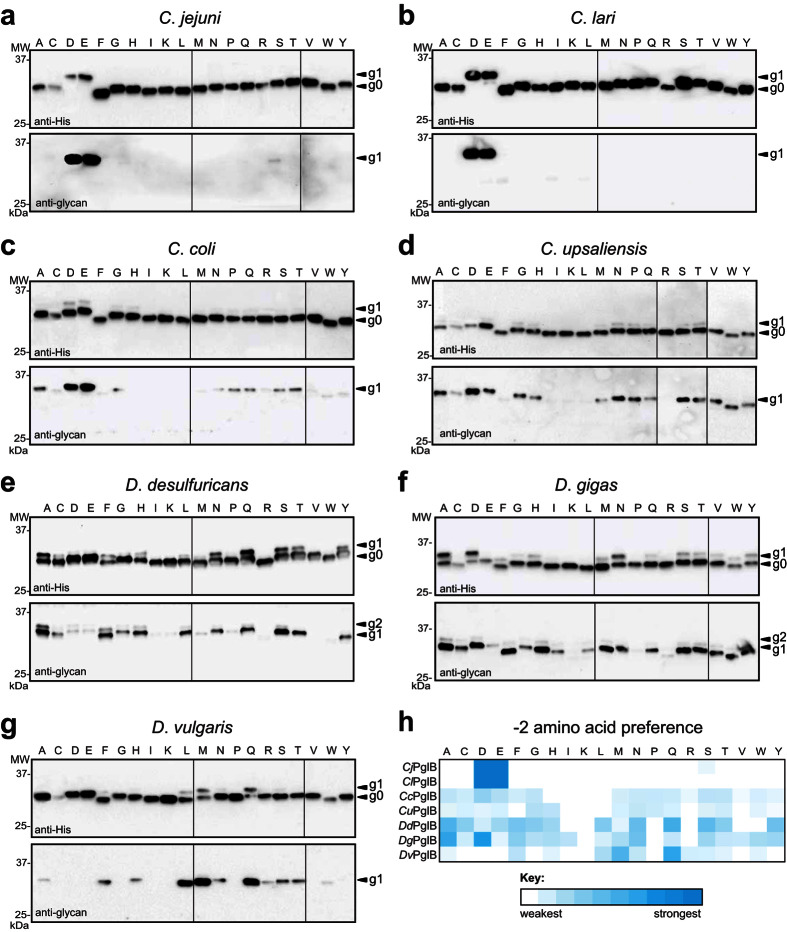
Figure 2. PglB homologs exhibit relaxed but different specificities for the −2 sequon residue.
(a–g) Immunoblot analysis of TCA-precipitated periplasmic fractions from CLM24 cells complemented with one of the PglB homologs indicated, and co-expressing the biosynthetic pathway for the C. jejuni heptasaccharide and a panel of scFv13-R4XQNAT variants (where X = one of the 20 amino acids). Blots were probed with anti-His antibodies against the C-terminal polyhistidine tag on acceptor protein or anti-glycan hR6 serum reactive with the C. jejuni heptasaccharide. The −2 residue in each acceptor protein is indicated across the top of each lane. The g0, g1, and g2 arrows indicate un-, mono-, and diglycosylated acceptor proteins, respectively. Molecular weight (MW) markers are indicated on the left. Due to the number of samples analyzed, all immunoblots shown are composites, delineated by dividing lines, from similar exposures. (h) Heatmap analysis of the relative −2 amino acid preference of each OST in (a–g). Relative preferences (weaker = white; stronger = dark cyan) were determined based on densitometric quantification of the percent glycosylated (defined as g1/g0 ratio) for each acceptor protein in the anti-His immunoblot.