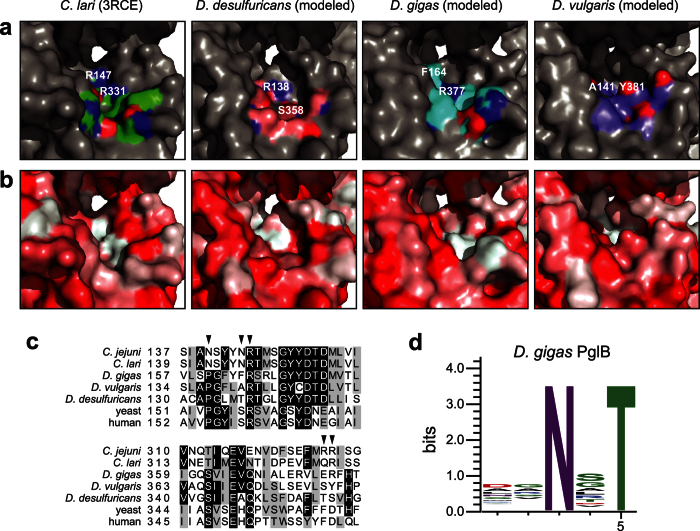
Figure 3. Molecular determinants of relaxed acceptor-site specificity.
(a,b) Homology models of DdPglB, DgPglB, and DvPglB compared to the crystal structure of ClPglB (pdb code: 3RCE). Differences in the presence and distribution of polar and electrostatic (a) and hydrophobic surfaces (b) are colored. Backbone residues are colored in green (C. lari), pink (D. desulfuricans), cyan (D. gigas), and purple (D. vulgaris). Red and blue shading indicates oxygen and nitrogen atoms, respectively. Conserved ClPglB residue R331 and corresponding residues in DdPglB (S358), DgPglB (R377) and DvPglB (Y381) are labeled. Conserved ClPglB residue R147 and its corresponding residue in DdPglB (R138) are labeled, but the homologous residues in DgPglB (R165) and DvPglB (R142) are not visible in the pocket. Adjacent residues F164 and A141 in DgPglB and DvPglB, respectively, are labeled. (c) Multiple sequence alignment of 5 PglB homologs compared to 2 eukaryotic STT3 sequences using Clustal Omega. Sequences span regions homologous to ClPglB residues 139–160 and 313–334. Shading indicates residue conservation. Some residues of interest are indicated with arrows across the top. (d) Sequence logo showing experimentally determined acceptor-site specificity of DgPglB using glycoSNAP-based library screening of YebF-R4Δ1-198XXNXT.