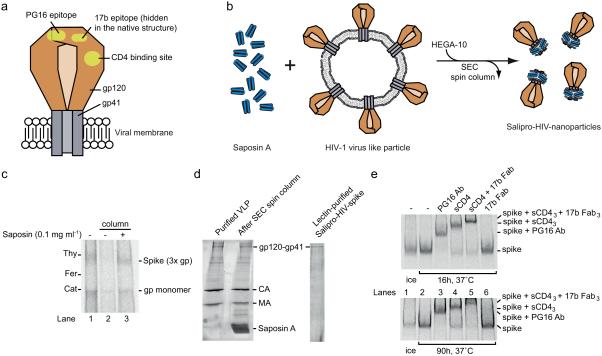
Figure 5. Stabilization of soluble and functional HIV-1 spike proteins within Salipro nanoparticles.
(a) Schematic illustration of the HIV-1 spike protein in the viral membrane. The HIV-1 Envelope glycoprotein, i.e. the spike, consists of two subunits, the peripheral gp120 subunit (brown) and the transmembrane gp41 subunit (grey), forming a heterotrimer. The binding site for the CD4 receptor and epitopes for the antibodies PG16 and 17b are shown (yellow). (b) Schematic illustration of the Salipro-HIV-spike reconstitution. Purified VLPs containing HIV-1 spikes (brown) were mixed with Saposin A (blue) and solubilized with HEGA-10 followed by detergent removal, leading to the formation of Salipro-HIV-spikes. (c) Reconstitution of the HIV-1 spike into Salipro nanoparticles. BN-PAGE analysis reveals that the solubility of the HIV-1 spike (3x gp) can be maintained by Saposin A upon detergent removal. (d) Lectin purification of Salipro-HIV-spikes. Particle purity was assayed using SDS-PAGE and Sypro Ruby protein staining. The gels show efficient removal of contaminating viral internal proteins. (e) The HIV-1 spike has a native fold and preserves its function in Salipro nanoparticles. The Salipro-HIV-spike particles were incubated at 37°C for 16 and 90 h followed by 2 h incubation at 37°C with 10 μg/ml of the HIV-1 spike ligands, PG16 antibody (150 kDa) (lane 3), sCD4 (50 kDa) (lane 4), sCD4 and 17b Fab (50 kDa) together (lane 5), 17b Fab alone (lane 6) or without a ligand (lane 2) and analyzed by BN-PAGE. A control sample without any ligand was kept on ice (lane 1). Binding of the ligands can be followed by the reduced migration of the Salipro-HIV-spike particles.