Figure 2. Msi1 induction expands the progenitor cell compartment and drives APC loss and RNA metabolism gene expression programs.
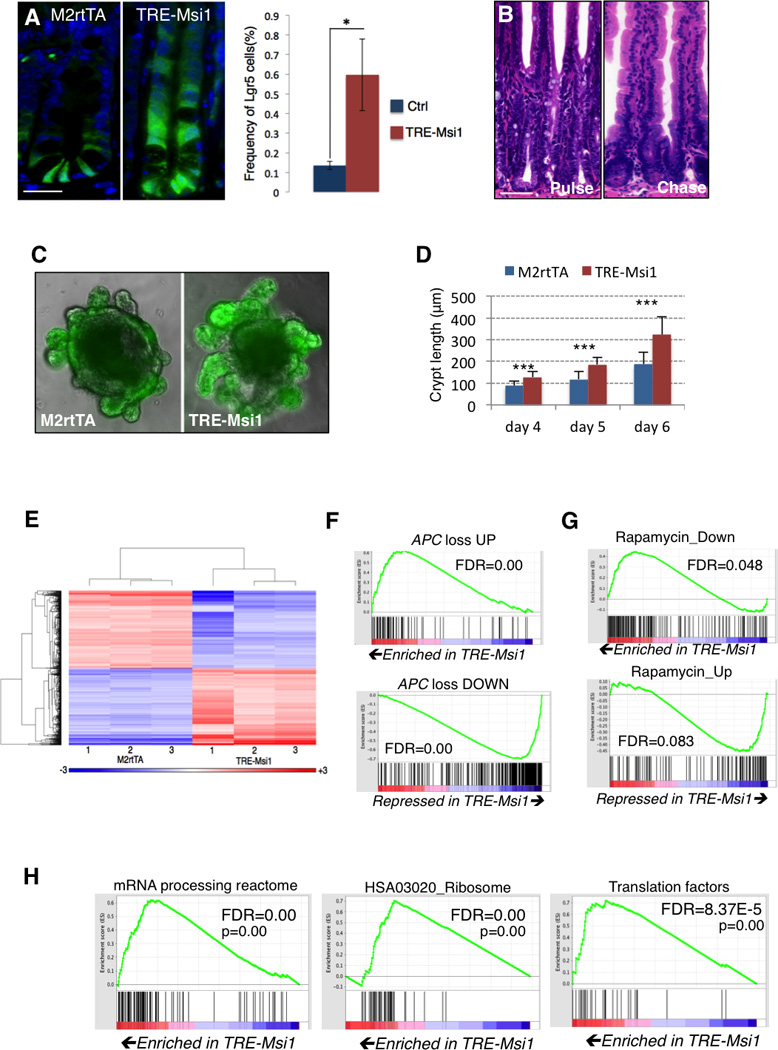
A. Msi1 induction in TRE-Msi1::Lgr5- eGFP-CreER mice results in an upward expansion of Lgr5-eGFP+ cells and an increase in the absolute frequency of Lgr5-eGFP+ cells, quantified by flow cytometry (right, n=3 mice per group, *p<0.05, Student’s t-test) (scale=50µm). B. TRE-Msi1 epithelium transformed by Dox induction for 48hrs revert to a phenotypically normal state persisting 2 months after Dox withdrawal (scale=100µm). C–D. In vitro culture of intestinal organoids derived from TREMsi1:: Lgr5-eGFP-CreER crypts followed by Dox induction in vitro. Crypt bud length is quantified in D (***: p<0.0005, Student’s t-test). E. Heatmap and hierarchical clustering of transcriptome profiles performed on the intestinal epithelium of 3 control (M2rtTA) and 3 TREMsi1 mice treated with Dox for 24 hours. F–H. Gene Set Enrichment Analysis (GSEA) of the TRE-Msi1 transcriptome identifies activation of genes induced by acute APC deletion in the intestinal epithelium (APC loss up) and suppression of genes downregulated after APC deletion (APC loss down) (F), along with an anti-correlation between the Msi1-induced transcriptome profile and the Peng_Rapamycin UP/DOWN gene sets (G), and an enrichment of expression of mRNA processing, ribosomal and translation factors upon Msi1 induction (H). FDR=False Discovery Rate. See also Supplemental Tables 1 and 2.
