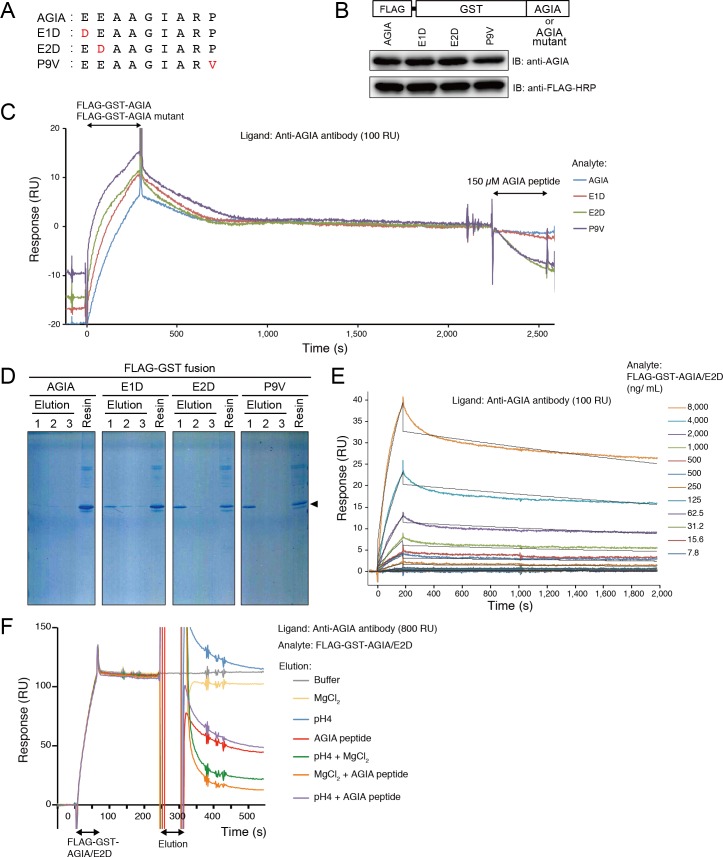
Fig 5. Development of AGIA/E2D tag.
(A) Amino acid substitution of AGIA tag sequence. Red characters indicate substituted residues. (B) Immunoblot of proteins fused with AGIA mutants. FLAG-GST-AGIA / AGIA mutants were subjected to western blotting. (C) Amino acid substitution in the AGIA tag enabled competitive dissociation. Anti-AGIA antibody was captured on a protein G-immobilized Biacore sensorchip at 100 RU. FLAG-GST-AGIA/AGIA mutant was injected for 300 sec, and dissociation was observed for 1,800 sec. Then, 150 μM AGIA peptide was injected for 300 sec. Blue, wild-type AGIA: red, AGIA/E1D; green, AGIA/E2D; purple, AGIA/P9V. (D) Competitive elution of AGIA mutants by AGIA peptide. Six hundred μL of cell-free synthesized FLAG-GST-AGIA/mutant was mixed with 20 μL anti-AGIA sepharose. After washing with 200 μL HBS three times, proteins were eluted 3 times by 200 μL of 150 μM AGIA peptide (Elution). After elution, the resin was boiled in SDS-PAGE sample buffer for 10 min (Resin). Fractions were applied to SDS-PAGE and CBB staining. (E) Kinetics assay of AGIA/E2D tag. Anti-AGIA antibody was captured on a protein G-immobilized Biacore sensorchip at 100 RU. Purified FLAG-GST-AGIA/E2D protein was injected for 180 sec as analyte. Black lines represent a global fit of a 1:1 interaction model to each kinetic data set. (F) Elution condition of AGIA/E2D tag. Anti-AGIA antibody was captured on a protein G immobilized Biacore sensorchip at 800 RU, and FLAG-GST-AGIA/E2D was injected for 60 sec. The sensorchip was then treated by following elution solutions; gray, HBS-EP+ buffer; yellow, 2M MgCl2; blue, 100 mM sodium acetate buffer pH4.0, red, 150 μM AGIA peptide; green, 100 mM sodium acetate buffer pH4.0 and 2M MgCl2; orange, 2M MgCl2 and 150 μM AGIA peptide; purple, 100 mM sodium acetate buffer pH4.0 and 150 μM AGIA peptide.