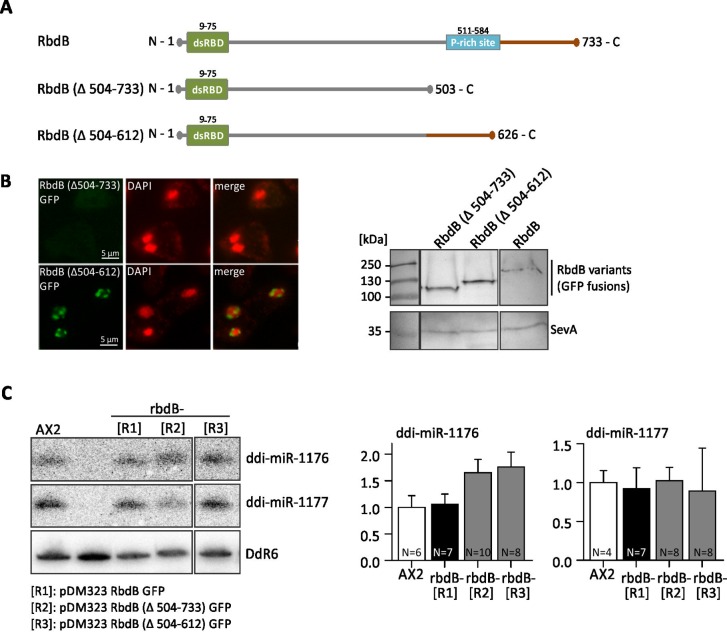
Fig 9. Analysis of truncated RbdB variants.
A: Schematic representation of RbdB and truncated protein variants. In RbdB Δ504–733 GFP, 230 amino acids were deleted from the C-terminus. RbdB Δ504–612 GFP lacks the Prich-site. B: left: Both truncated RbdB-GFP versions were expressed in the knockout background and visualized by fluorescence microscopy. RbdB Δ504–612 GFP showed the same distribution as RbdB GFP. In contrast, RbdB Δ504–733 GFP was not detectably by fluorescence microscopy. right: Western Blot showing expression of RbdB Δ504–733 GFP (84 kDa), RbdB Δ504–612 (98 kDa) and of RbdB GFP (109 kDa). Note that all proteins run at higher molecular levels than calculated. SevA (40 kDa) is shown as loading control. C: left: Northern Blot analysis of rbdB- strains expressing RbdB GFP [R1], RbdB Δ504–733 GFP [R2], RbdB Δ504–612 GFP [R3] on miRNAs. 12 μg total RNA were loaded per lane. As a control, RNA from the AX2 wt and from an rbdB- strain was used. The mature miRNAs ddi-miR-1176 and ddi-miR-1177 were detected by 32P labelled probes as described in Fig 3A. Hybridisation to snoRNA DdR6 was used as a loading control. Right: miRNA signals (ddi-miR-1176 left, ddi-miR-1177 right) were quantified relative to DdR6 from different Northern Blots and normalized to the AX2 wt. R1-R3: rbdB- mutants were transformed with pDM323 RbdB Δ504–733 GFP (R1), pDM323 RbdB Δ504–612 GFP (R2) and with pDM323 RbdB GFP (R3). According to paired t-test, no significant difference was seen in miRNA accumulation between the wild type and the mutants.