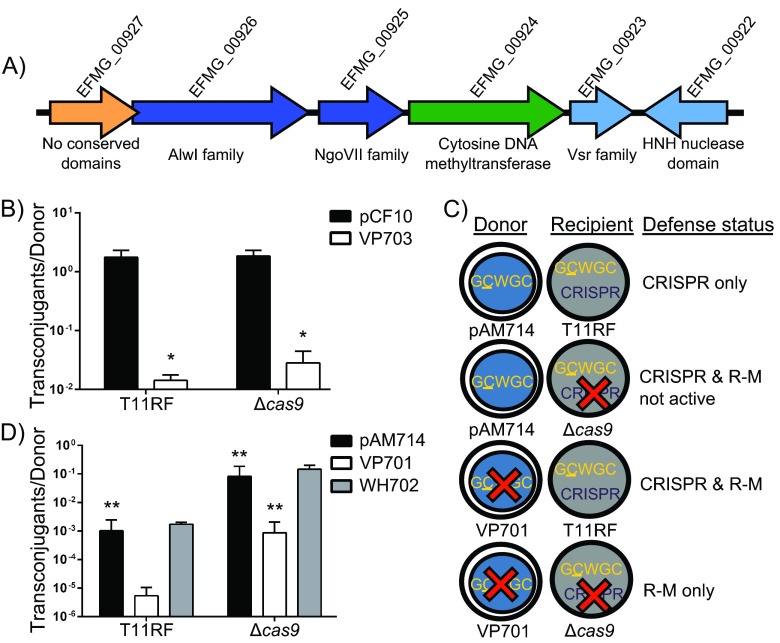
FIG 3 .
CRISPR-Cas and R-M provide additive defense against PRPs in E. faecalis. (A) Organization of the predicted R-M locus of T11; multiple predicted REases are encoded near the MTase. (B) Conjugation frequencies with T11RF and T11RFΔcas9 strains as recipients in mating reactions with OG1SSp pCF10 and VP703 as donors. P values are relative to transfer of OG1SSp pCF10 to T11RF: *, P < 0.05. (C) Schematic representing donor and recipient strains used to assess the individual and collective contributions of R-M and CRISPR-Cas to genome defense. (D) Conjugation frequencies with T11RF and T11RFΔcas9 strains as recipients (x axis) and with OG1SSp pAM714 (black columns), OG1SSp pAM714 ΔEfaRFI (VP701; white columns), and OG1SSp pAM714 ΔEfaRFI + EfaRFI (WH702; gray columns) as donors. Frequencies are shown as the ratios of transconjugants to donors. Results of these experiments show that the combined effects of CRISPR-Cas and R-M outweigh the effect of either system alone. Data represent results of a minimum of three independent conjugations for all experiments shown. P values are relative to transfer of pAM714 from VP701 to T11RF: **, P < 0.005. Significance in the data in panels B and D was assessed using a one-tailed Student’s t test.