Figure 2.
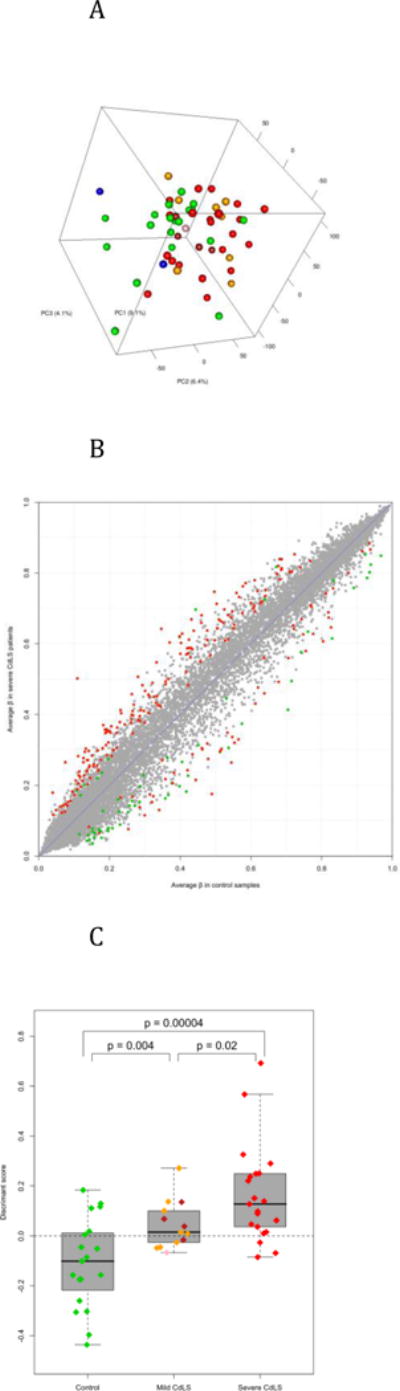
Differential methylation. A) Principal components analysis of LCLs using all measured CpG sites on autosomes. Colors indicate disease status (green = control; red = severe CdLS; orange = mild CdLS with NIPBL mutation; brown = CdLS with SMC1A mutation; pink = CdLS with SMC3 mutation; and blue = Roberts Syndrome). B) Differential methylation between control and severe CdLS samples. Each dot represents one autosomal site. Significant sites (p < 0.01 and | Δβ | > 0.05) were highlighted (green = CGI sites and red = non-CGI sites). C) The control, mild CdLS, and severe CdLS samples could be distinguished according to their methylation pattern. The y-axis indicates the discriminant score that is corresponding to the relative similarity of each sample to the centroids of control and severe CdLS groups. The scores of controls and severe patients were obtained via a leave-one-out procedure, and the scores of mild patients were based on the 283 sites differentially methylated between controls and severe patients (details in Methods). By default, samples with score > 0 would be classified as CdLS. Each diamond represents a sample (colored as in Figure 2A). The p values are the results of Student’s t test.
