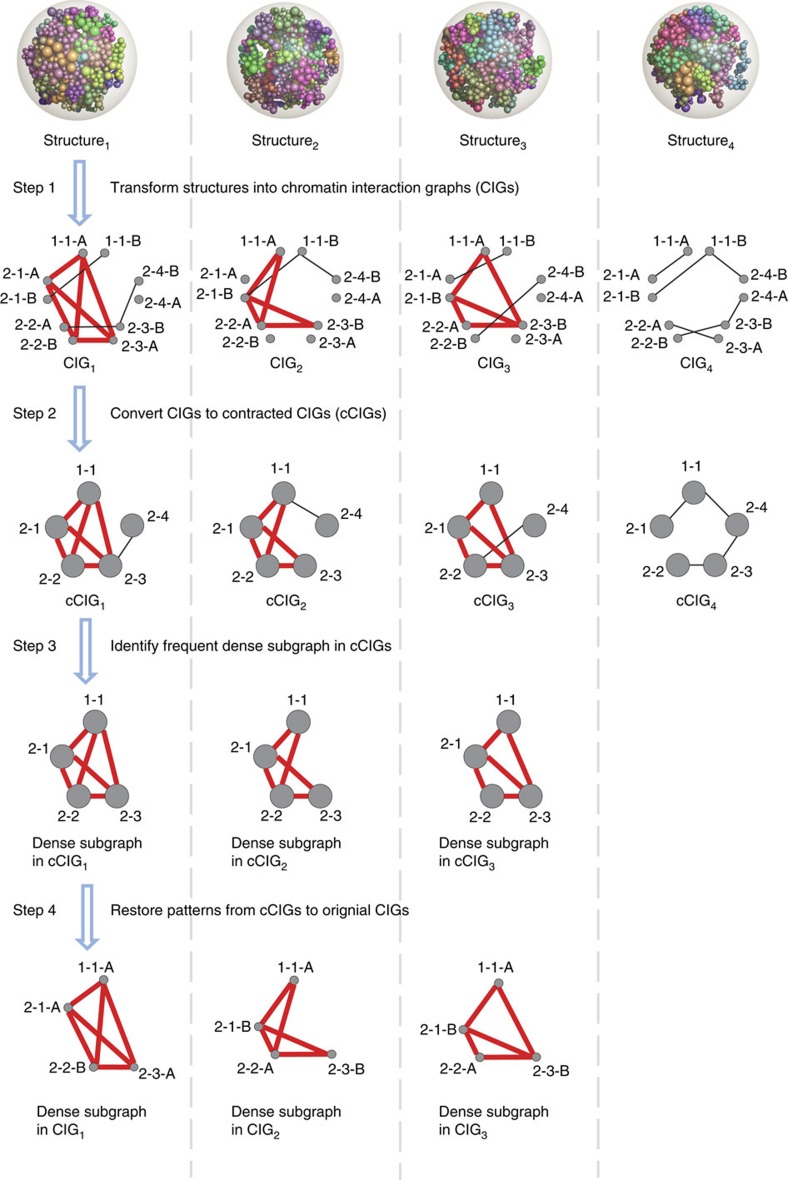
Figure 1. The overall procedure to discover frequent dense clusters.
Each 3D genome structure is transformed into a CIG where a node represents a domain and an edge represents a contact between two domains. In this example, 4 CIGs with 10 nodes are built. Each node is labelled by the triplet L1–L2–L3, where L1 indicates the chromosome index, L2 indicates the domain index among all domains in its chromosome and L3 (a letter A or B) indicates which copy of the chromosome the domain comes from. For example, nodes 2-3-A and 2-3-B indicate ‘twin' nodes are from the 3rd domain of chromosome 2, in its two homologous copies. Step 1: Each genome structure is transformed into a CIG. Step 2: in each CIG, we merge any two ‘twin' nodes that represent homologous domains. This step yields a collection of four contracted graphs without isomorphism (termed cCIG, where each node is a merged domain, labelled by L1 and L2). Step 3: We identify the dense subgraphs that frequently occur across many networks using a tensor-based computational method. Step 4: We restore each frequent dense subgraph to its un-contracted form in the original CIGs, and after mining on these subgraphs with ‘coupled isomorphism' we identify the final set of frequent dense subgraphs.