FIG 6 .
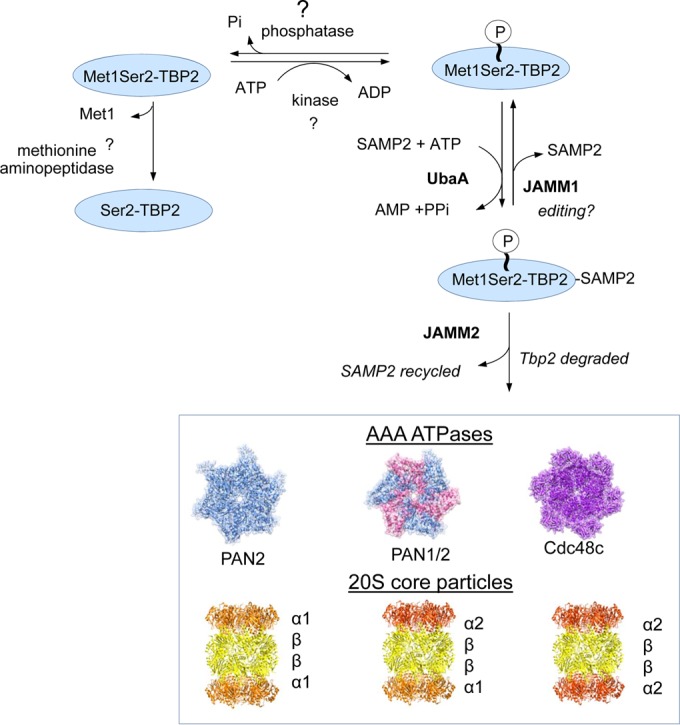
Model of regulated turnover of TBP2 by the archaeal SAMP proteasome system (SPS). In this model, the phosphorylation status of TBP2 Ser2 is an important factor that regulates TBP2 turnover by the archaeal SPS. The kinase and phosphatase enzymes that control phosphorylation of TBP2 Ser2 and the methionine aminopeptidase that cleaves TBP2 Met1 have yet to be identified. However, evidence suggests that the addition of a phosphoryl group to Ser2 inhibits the ability of methionine aminopeptidase to remove Met1 from TBP2. The Met1-Ser2(p) form of TBP2 is thought to be susceptible to sampylation by the E1-like UbaA and destruction by the archaeal SPS. The proteasomal AAA ATPases (Rpt-like PAN2 and Cdc48c) are used to recognize, unfold, and translocate TBP2 into the proteolytic chamber of the CPs for destruction by the proteasome system, while JAMM2 would remove and recycle the SAMP2 moiety from the TBP2 substrate during this process. JAMM1 is a desampylase that independently cleaves SAMPs from target proteins and, thus, may serve as a proofreading enzyme to ensure proper substrate recognition by the archaeal SPS.
