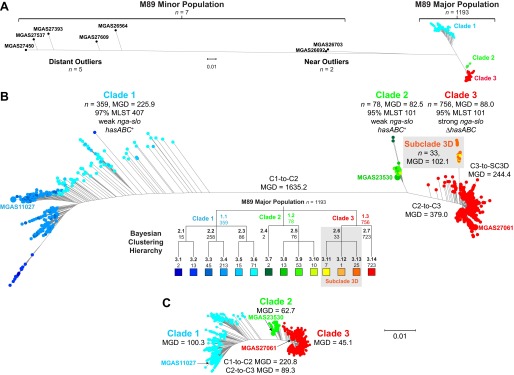
FIG 2 .
Genetic relationships among emm89 strains. Genetic relationships were inferred by the neighbor-joining method based on concatenated core chromosomal SNP data using SplitsTree. (A) Genetic relationships based on 28,425 SNPs identified among the members of the entire population of 1,200 emm89 strains. (B) Genetic relationships based on 11,846 SNPs identified among the 1,193 major population strains. Isolates are colored by cluster as determined using Bayesian analysis of population structure (BAPS) as indicated in the hierarchy below the figure. Three major clades (C1, C2, and C3) are defined at the first level of clustering. Subclade 3D (SC3D), a recently emerged and expanding population of strains in Finland, is defined at the second level of clustering. Indicated for the inferred phylogenies are the mean genetic distances (MGDs), both inter- and intraclade, measured as differences in core chromosomal SNPs. The mean genetic distance among strains within clades is less than the MGD to strains of the nearest neighboring clade(s). Bootstrap analysis with 100 iterations gives 100% confidence for all of the clade-to-clade branches (i.e., C1-C2, C2-C3, and C3-SC3D). (C) Genetic relationships based on 8,989 SNPs identified among the major population of 1,193 strains, filtered to exclude horizontally acquired sites as inferred using Gubbins. Exclusion of sites attributed to horizontal gene transfer events collapses the MGD strain-to-strain both within and between the clades. The MGD within the clades remains less than the MGD to the nearest neighboring clade(s). Trees in panels B and C are illustrated at the same scale.