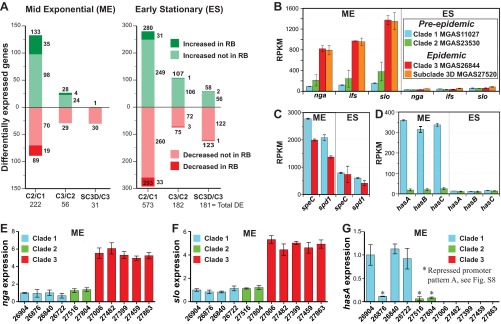
FIG 7 .
RNAseq and qPCR expression analyses. (A) Genes significantly altered in transcript level at 1.5-fold change or greater in RNAseq. The numbers shown refer to the total number of differentially expressed genes for each comparison. The representative strains of each clade analyzed are C2/C1 (MGAS23530/MGAS11027), C3/C2 (MGAS26844/MGAS23530), and SC-3D/C3 (MGAS27520/MGAS26844). (B) Transcript levels for the nga-ifs-slo operon. The transcript levels of nga, ifs, and slo were significantly (4- to 8-fold) (P < 0.05) greater in the epidemic strains than in the preepidemic strains. The index in panel B applies to panels B, C, and D. (C) Transcript levels of speC and spd1. Transcript levels of speC and spd1 were significantly greater in the preepidemic strain at the exponential growth phase (P < 0.01). (D) Transcript levels for the hasABC operon. Transcription of hasABC was very weak for the clade 2 strain at both growth phases and was significantly less than for the clade 1 strain at the mid-exponential growth phase (P < 0.002). (E, F, and G) Relative transcript levels as measured by qPCR for nga, slo, and hasA, respectively. Differences in strain-to-strain expression were assessed by one-way ANOVA. Expression of nga and slo was significantly greater for all 5 clade 3 strains than for all 6 clade 1 and 2 strains (P < 0.001). All 3 clade 1 and 2 strains with hasABC weak/repressed promoter pattern A expressed significantly less hasA than all 3 clade 1 strains with strong/derepressed promoter pattern B (P < 0.001). Levels of expression of hasA were not significantly different among all 3 strains with weak/repressed promoter pattern A and all 5 genetically acapsular clade 3 strains. RB, recombination block; RPKM, reads per kilobase per million reads mapped.