ABSTRACT
The future sustainable production of chemicals and fuels from nonpetrochemical resources and reduction of greenhouse gas emissions are two of the greatest societal challenges. Gas fermentation, which utilizes the ability of acetogenic bacteria such as Clostridium autoethanogenum to grow and convert CO2 and CO into low-carbon fuels and chemicals, could potentially provide solutions to both. Acetogens fix these single-carbon gases via the Wood-Ljungdahl pathway. Two enzyme activities are predicted to be essential to the pathway: carbon monoxide dehydrogenase (CODH), which catalyzes the reversible oxidation of CO to CO2, and acetyl coenzyme A (acetyl-CoA) synthase (ACS), which combines with CODH to form a CODH/ACS complex for acetyl-CoA fixation. Despite their pivotal role in carbon fixation, their functions have not been confirmed in vivo. By genetically manipulating all three CODH isogenes (acsA, cooS1, and cooS2) of C. autoethanogenum, we highlighted the functional redundancies of CODH by demonstrating that cooS1 and cooS2 are dispensable for autotrophy. Unexpectedly, the cooS1 inactivation strain showed a significantly reduced lag phase and a higher growth rate than the wild type on H2 and CO2. During heterotrophic growth on fructose, the acsA inactivation strain exhibited 61% reduced biomass and the abolishment of acetate production (a hallmark of acetogens), in favor of ethanol, lactate, and 2,3-butanediol production. A translational readthrough event was discovered in the uniquely truncated (compared to those of other acetogens) C. autoethanogenum acsA gene. Insights gained from studying the function of CODH enhance the overall understanding of autotrophy and can be used for optimization of biotechnological production of ethanol and other commodities via gas fermentation.
IMPORTANCE
Gas fermentation is an emerging technology that converts the greenhouse gases CO2 and CO in industrial waste gases and gasified biomass into fuels and chemical commodities. Acetogenic bacteria such as Clostridium autoethanogenum are central to this bioprocess, but the molecular and genetic characterization of this microorganism is currently lacking. By targeting all three of the isogenes encoding carbon monoxide dehydrogenase (CODH) in C. autoethanogenum, we identified the most important CODH isogene for carbon fixation and demonstrated that genetic inactivation of CODH could improve autotrophic growth. This study shows that disabling of the Wood-Ljungdahl pathway via the inactivation of acsA (encodes CODH) significantly impairs heterotrophic growth and alters the product profile by abolishing acetate production. Moreover, we discovered a previously undescribed mechanism for controlling the production of this enzyme. This study provides valuable insights into the acetogenic pathway and can be used for the development of more efficient and productive strains for gas fermentation.
INTRODUCTION
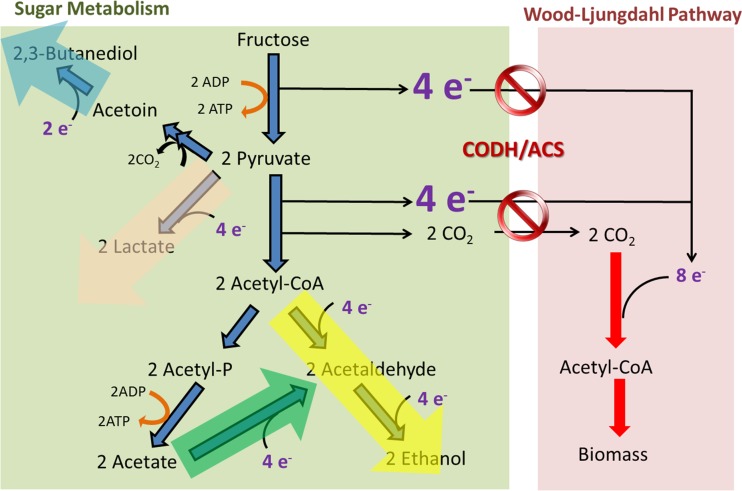
Acetogenic bacteria employ the Wood-Ljungdahl pathway (WLP) to fix CO2 (in the presence of H2) and CO into the central metabolite acetyl coenzyme A (acetyl-CoA). It is the only linear CO2 fixation pathway known and the most thermodynamically efficient pathway in acetate synthesis (1). As a consequence, the WLP is a prime candidate for the earliest autotrophic pathway in the origin of life (2). Terrestrial production of acetate by acetogens is estimated to be at least 1013 kg/annum, accounting for more than 20% of the fixed carbon on Earth, highlighting their significant role in the global carbon cycle (3). The ability to fix C1 gases also makes acetogens attractive process organisms for the production of chemicals and fuels. Fermentation processes that recycle waste gases from industrial processes or syngas generated from any biomass source are on the verge of commercialization (4) and offer significant greenhouse gas emission savings (5) to meet the climate goals under the Paris Agreement (6).
Crucial to the function of the WLP are the enzymes carbon monoxide dehydrogenase (CODH) and acetyl-CoA synthase (ACS). CODH catalyzes the interconversion of CO and CO2 according to the equation CO + H2O ←→ CO2 + 2H+ + 2e−. CO is a potent electron donor (CO2/CO reduction potential of −558 mV [pH 7.0]) (7). Relatively few acetogens are, however, able to grow on CO alone because of growth inhibition resulting from the sensitivity of metal-containing enzymes to CO (8). CODH can also form a bifunctional complex with the ACS that couples the reduction of CO2 and the formation of acetyl-CoA. This unique enzyme has been extensively studied at the protein level (9–11), but in vivo and genetic studies of CODH in acetogens are lacking.
Clostridium autoethanogenum, a model acetogen, is able to grow on CO as a sole carbon and energy source and produce ethanol, acetate, 2,3-butanediol, and lactate (12, 13). Whole-genome sequencing of this acetogen revealed the presence of three putative CODHs: CAETHG_1620-1621 (acsA), CAETHG_3005 (cooS1), and CAETHG_3899 (cooS2) (14). acsA is in an 18-kbp cluster with genes of the WLP and is predicted to encode the CODH component of the CODH/ACS complex, while cooS1 is localized upstream of a gene encoding a putative 4Fe-4S ferredoxin binding domain-containing protein and ferredoxin oxidoreductase. cooS2 appears to be an orphan. Transcriptomic studies have shown that all three genes are expressed, with acsA and cooS1 being among the most highly expressed genes within the genome (15, 16).
Given the number of CODHs within C. autoethanogenum, it is unknown whether all of them are essential for autotrophic growth and are true isozymes or have distinct functions. Here, we addressed this question by independently inactivating all three CODH-encoding genes by ClosTron mutagenesis (17–19) and then examined the impact on autotrophy and product formation. The mutant strain in which acsA was inactivated is particularly interesting because it essentially has a disabled WLP and, as a consequence, displays a radically different metabolite distribution, including the complete abolition of acetate formation.
Intriguingly, compared to the acsA genes of other acetogens, that of C. autoethanogenum uniquely contains an in-frame TGA stop codon. The encoded enzyme is therefore predicted to be truncated by some 231 amino acids and the gene effectively split in two: CAETHG_1621 and CAETHG_1620. By using FLAG-tagged acsA variants, the apparent acsA truncation event was investigated.
RESULTS
acsA is essential for autotrophy in C. autoethanogenum.
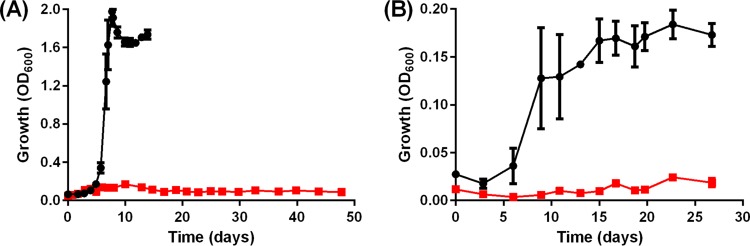
To determine the roles of acsA in supporting autotrophy in C. autoethanogenum, we first inactivated the gene by ClosTron mutagenesis (17–19), which resulted in an acsA knockout (KO) strain (see Fig. S1 in the supplemental material). Both the KO and wild-type (WT) strains were subjected to autotrophic batch growth on either CO or H2-CO2 (2:1) to assess the role of acsA in catalyzing CO oxidation and/or fixation of CO2 (using H2 as a reductant). As shown in Fig. 1A, the acsA KO strain displayed no sign of growth on CO following 48 days of incubation, whereas the WT reached an optical density at 600 nm (OD600) of 1.98 after day 8. Under H2-CO2 conditions, the acsA KO strain was unable to grow within 27 days, whereas the WT achieved stationary phase (OD600 of 0.17) after ~day 15 (Fig. 1B). In an attempt to restore autotrophy, plasmid pMTL83151-PacsA-acsAfull containing acsA was conjugated into the acsA KO strain to generate a complementation strain. The complementation strain was able to restore growth and acetate formation on CO, albeit growing to an OD600 of 1.10 after a growth lag phase of ~21 days (see Fig. S2 in the supplemental material) and generating 15% less acetate (P = 0.009). The growth characteristics of the plasmid control strain (harboring pMTL83151-PacsA) were generally similar to those of the WT, except that the lag phase was longer by 5 days (Fig. 1A; see also Fig. S2 in the supplemental material).
FIG 1 .
The acsA KO strain is unable to grow autotrophically on 200 kPa CO (A) or 130 kPa H2 plus 70 kPa CO2 (B). Symbols: black circles, WT (n = 4 for CO; n = 3 for H2-CO2); red squares, acsA KO strain (n = 3). Error bars show the standard error of the mean.
Inactivation of acsA abolishes acetate formation during heterotrophic growth.
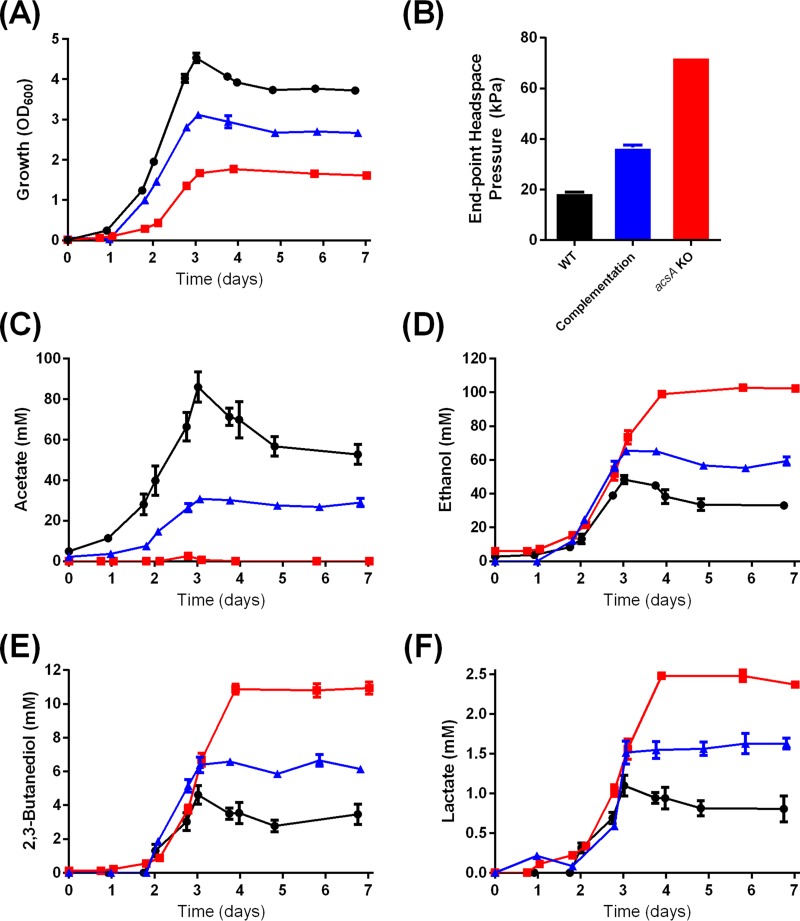
In order to gain an insight into the role of acsA during heterotrophic growth, we next investigated the growth and product profile of the acsA KO strain on fructose. The acsA KO strain fully exhausted the supplemented 10 g/liter fructose but reached a 61% lower OD600 (P < 0.0001) and exhibited a longer growth lag phase than the WT (Fig. 2A). The 3.8-fold higher headspace pressure recorded at the end of the growth experiment with the acsA KO strain (Fig. 2B) indicates that more CO2 was being released from fructose metabolism as a result of the organism’s inability to reassimilate the released CO2 as a consequence of its dysfunctional WLP.
FIG 2 .
Growth, headspace pressure, and metabolite profiles of the C. autoethanogenum WT, acsA KO, and complementation strains on 10 g/liter fructose. Panels: A, growth profile; B, headspace pressure profile; C, acetate profile; D, ethanol profile; E, 2,3-butanediol profile; F, lactate profile. Black, WT; red, acsA KO strain; blue, complementation strain. n = 3. Error bars show the standard error of the mean.
In terms of metabolite production, the acsA KO strain only transiently produced a trace amount of acetate (2.6 mM on day 2.8) while growing on fructose (Fig. 2C). In contrast, the WT strain generated 86.0 mM acetate under the same conditions (Fig. 2C). In the acsA KO strain, most of the carbon from fructose was diverted from acetate toward reduced products ethanol and 2,3-butanediol and toward lactate, as evident in increases of 113, 138, and 125%, respectively, relative to the WT (Fig. 2D to F). Similar to autotrophic growth on CO (mentioned earlier), the plasmid expression of acsAfull in the acsA KO strain partially restored the phenotypes of heterotrophic growth by reducing the growth lag phase to WT levels and increased the OD600 from 1.77 (acsA KO) to 3.11, which is 69% of the WT level (Fig. 2A). The complementation strain also synthesized 30.78 mM acetate (up from the 2.5 mM of the acsA KO), 65.35 mM ethanol (down from the 102.74 mM of the acsA KO), and 6.66 mM 2,3-butanediol (down from 10.95 mM) (Fig. 2C to E). Plasmid expression of acsA (pMTL83151-PacsA-acsAfull) in the WT had only minimal effects on growth and metabolite production during growth on CO (see Fig. S3 in the supplemental material) or under heterotrophic growth conditions (see Fig. S4 in the supplemental material).
Translational readthrough of acsA.
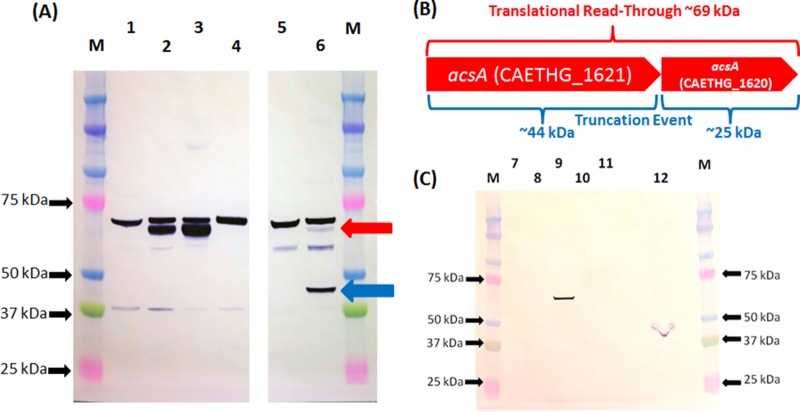
Genome sequencing of C. autoethanogenum (14, 20, 21) revealed the presence of an internal TGA stop codon within the acsA gene, splitting the gene into coding sequences (CDSs) CAETHG_1621 (1,203 bp) and CAETHG_1620 (684 bp) (see Fig. S5 in the supplemental material). Sanger sequencing confirmed the presence of the TGA stop codon (data not shown).
By fusing a FLAG tag to the N or C terminus of AcsA, the acsA translation pattern in C. autoethanogenum and Escherichia coli was investigated by Western blot analysis. In addition, the TGA stop codon was replaced with a TCA or TAA codon by splicing by overhang extension PCR (SOE-PCR). Modified genes were expressed from plasmids. The truncation of the 69-kDa full-length AcsA protein can result in proteins of 44 and/or 25 kDa (Fig. 3B). With a C-terminal FLAG tag, a 69-kDa protein was detected in C. autoethanogenum crude lysates (Fig. 3A), while a 25-kDa protein band was absent. With the N-terminal FLAG tag variant, the 44- and 69-kDa proteins were both detected, with the intensity of the 44-kDa protein band being higher. When the TGA codon was replaced with a TCA codon, only the 69-kDa protein was detected. These results indicate that translational readthrough of the acsA TGA codon occurs with low frequency in C. autoethanogenum and there is no independent translation of the downstream CDS.
FIG 3 .
Examination of AcsA translation pattern with FLAG-tagged protein in C. autoethanogenum and E. coli. (A) Western blot analysis of C. autoethanogenum transconjugant crude lysates. Lanes: M, Bio-Rad kaleidoscope Precision Plus protein ladder; 1, 13 µg of soluble lysate of pMTL8315-PacsA plasmid control; 2, 13 µg of soluble lysate of pMTL83151-PacsA-acsA(TGA)-FLAG; 3, 13 µg of soluble lysate of pMTL83151-PacsA-acsA(TCA)-FLAG; 4, 13 µg of soluble lysate of pMTL83151-PacsA-acsA(TAA)-FLAG; 5, 6.7 µg of insoluble lysate of pMTL83151-PacsA plasmid control; 6, 6.7 µg of insoluble lysate of pMTL83151-PacsA-FLAG-acsA(TGA). The red arrow indicates the position of a mature 69-kDa protein; the blue arrow indicates the position of the larger 44-kDa truncated protein. The ca. 70-kDa band present in all crude lysates from C. autoethanogenum is a consequence of nonspecific binding of the anti-FLAG antibody to a native C. autoethanogenum protein, most likely DnaK (encoded by CAETHG_2891), which shares 7/8 amino acid identity with FLAG and is of the appropriate predicted size. (B) Schematic showing the expected protein sizes of C. autoethanogenum AcsA in the event of translational readthrough or truncation. (C) Western blot analysis of E. coli transformant crude lysates. Lanes: M, Bio-Rad kaleidoscope Precision Plus protein ladder; 7, 15 µg of soluble lysate of pMTL83151-PacsA plasmid control; 8, 15 µg of soluble lysate of pMTL83151-PacsA-acsA(TGA)-FLAG; 9, 15 µg of soluble lysate of pMTL83151-PacsA-acsA(TCA)-FLAG; 10, 15 µg of soluble lysate of pMTL83151-PacsA-acsA(TAA)-FLAG; 11, 39 µg of soluble lysate of pMTL83151-PacsA-FLAG-acsA(TGA); 12, 38 µg of insoluble lysate of pMTL83151-PacsA-FLAG-acsA(TGA).
E. coli is a model microorganism for the study of selenocysteine synthesis and is capable of selenocysteine incorporation (22, 23). Lysates of E. coli cells that expressed acsA variants encoding either N- or C-terminally FLAG-tagged proteins failed to produce the 69-kDa full-size AcsA product (Fig. 3C). Instead, the 44-kDa truncated protein was detected. However, the acsA variant with a TCA serine codon successfully generated the full-size ~69-kDa protein (Fig. 3C). Replacement of the internal stop codon with a TAA stop codon completely eliminated the translation of the full-size 69-kDa AcsA peptide in both C. autoethanogenum and E. coli (Fig. 3A and C).
cooS1 and cooS2 are dispensable for autotrophy.
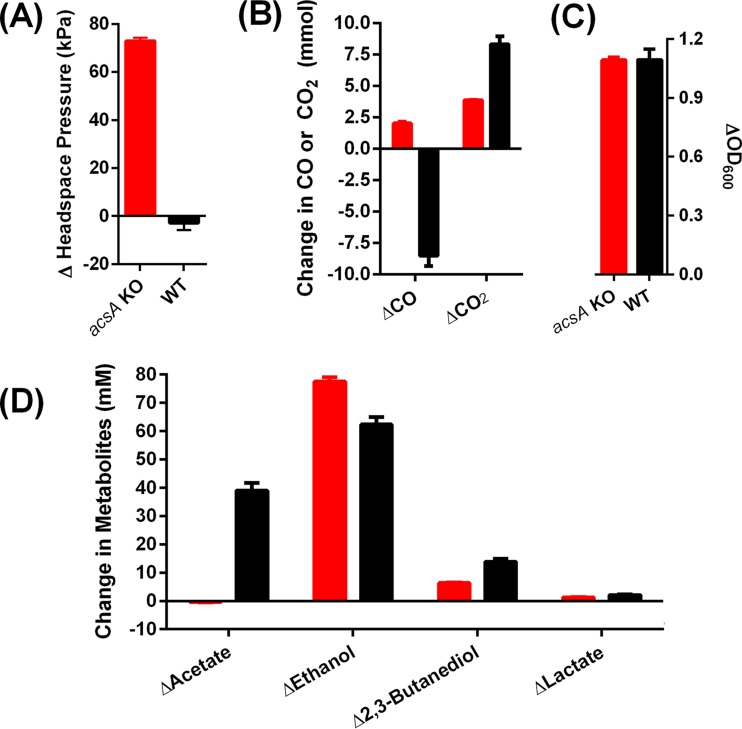
Besides acsA, the genome of C. autoethanogenum contains two additional putative CODHs: cooS1 and cooS2. These are unable to compensate for acsA and are therefore predicted to be monofunctional, as shown above. Accordingly, the acsA KO strain would be expected to grow on fructose (10 g/liter) and oxidize CO (22.4 mmol) into CO2 by using the unperturbed cooS1- and/or cooS2-encoded CODHs. In experiments that compared the acsA KO strain to the WT grown mixotrophically on CO plus fructose, the acsA KO strain generated 2.03 mmol of CO, while the WT control consumed 8.52 mmol of CO (Fig. 4B). Both strains completely exhausted the fructose at the end of the experiment (data not shown)
FIG 4 .
Changes in biomass, headspace, and metabolite levels between the start and finish of a mixotrophic-growth experiment with the acsA KO and WT strains on 10 g/liter fructose and 200 kPa CO. Panels: A, change in headspace pressure; B, change in headspace CO or CO2; C, change in growth based on OD600; D, change in metabolites. Columns: red, acsA KO strain; black, WT. n = 3. Error bars show the standard error of the mean.
In the absence of a WLP, the consumption of 10 g/liter fructose (or an absolute amount of 2.8 mmol) is expected to produce a maximum CO2 level of 5.6 mmol (1 mol of fructose yields 2 mol of pyruvate, which in turn is decarboxylated into acetyl-CoA with the concomitant release of 2 mol of CO2; Fig. 5). In acetogens, a large proportion of the released CO2 is fixed into acetyl-CoA. Since the WT produced 8.3 mmol of CO2 during mixotrophic growth on 2.8 mmol fructose and 8.52 mmol of CO (Fig. 4B), which is greater than the theoretical maximum of 5.6 mmol of CO2, CO oxidation must have occurred, as opposed to the direct use of CO in the carbonyl branch of the WLP. These results indicate that while the WT strain is able to oxidize CO during mixotrophic growth, neither cooS1 nor cooS2 in the acsA KO strain is able to catalyze CO oxidation. Instead, the physiological roles of these monofunctional CODHs under mixotrophic growth conditions may lie in the direction of CO2 reduction since additional CO was produced by the acsA KO strain.
FIG 5 .
Inactivation of CODH/ACS in C. autoethanogenum generates excess reducing equivalents that are consumed in biochemical reactions that lead to ethanol, 2,3-butanediol, and lactate formation.
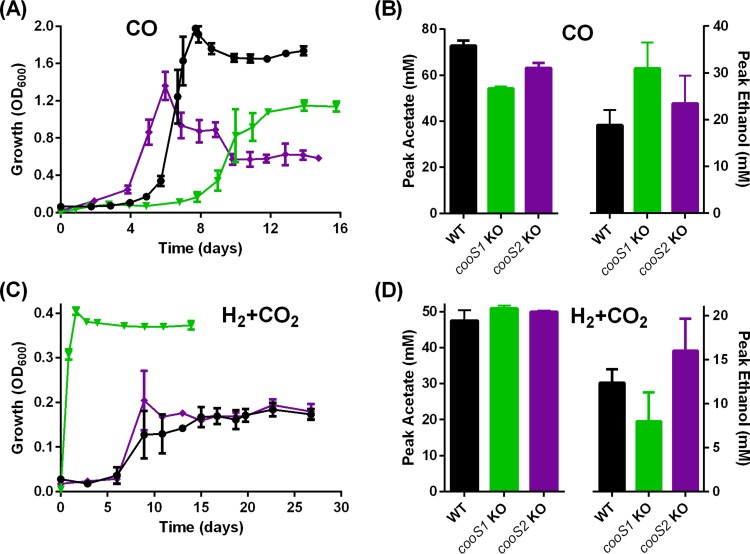
By ClosTron mutagenesis, cooS1 and cooS2 of C. autoethanogenum were independently inactivated to assess their roles in autotrophy (see Fig. S1 in the supplemental material). When grown on pure CO, the cooS1 KO strain displayed some growth deficiencies, including an ~2.9-day-longer lag phase, and achieved a 42% lower OD600 (P = 0.0002) than the WT (Fig. 6A). It produced 25% less acetate (P = 0.0004) and a similar amount of 2,3-butanediol (data not shown) but 64% more ethanol (not statistically significant) than the WT (Fig. 6B). The reduced growth of the cooS1 KO strain in the presence of CO may result in the accumulation of excess reducing equivalents that are consumed in ethanol biosynthetic pathways. Remarkably, under H2-CO2 conditions, the cooS1 KO strain is able to grow without an apparent lag phase and also reaches twice the OD600 of the WT (Fig. 6C).
FIG 6 .
Growth and metabolite profiles of the C. autoethanogenum WT and cooS1 and cooS2 KO strains on CO or H2-CO2. Panels: A, growth profile on CO; B, metabolite profile on CO; C, growth profile on H2-CO2; D, metabolite profile on H2-CO2. Columns: black, WT (n = 4 for CO, n = 3 for H2-CO2); green, cooS1 KO strain (n = 3); purple, cooS2 KO strain (n = 3). Error bars show the standard error of the mean.
Unlike the autotrophic growth of the cooS1 KO strain with CO, that of the cooS2 KO strain is not significantly affected. The cooS2 KO strain even showed a mild decrease in the growth lag phase but a lower final OD600 in the presence of CO than the WT (Fig. 6A). This result, combined with a lack of impact on H2-CO2 autotrophic growth (Fig. 6C), suggests that cooS2 is not heavily involved in autotrophy of C. autoethanogenum.
DISCUSSION
Understanding the fundamentals of C1 metabolism in acetogens is a prerequisite for their further development as a chassis for the sustainable production of chemicals and fuels from waste gases. By independently disrupting all three CODH isogenes in C. autoethanogenum, we investigated their roles in supporting autotrophy. The complete absence of growth of the acsA KO strain on CO or H2-CO2 demonstrated that acsA is absolutely essential for autotrophy under both CO and H2-CO2 conditions and that unperturbed cooS1 and cooS2 are unable to compensate for the loss of acsA function. In Methanosarcina acetivorans, which has two CODH/ACS paralogs, deletion studies showed that the microorganism can grow autotrophically in CO when one of these genes was deleted but not when both were deleted (24).
During glycolysis, 1 mol of hexose sugar is metabolized to 2 mol of acetyl-CoA, 2 mol of CO2 (generated during the pyruvate:ferredoxin oxidoreductase reaction), and 8 reducing equivalents (Fig. 5). Acetogens such as C. autoethanogenum utilize the 8 reducing equivalents to reassimilate the released 2 mol of CO2 into an additional acetyl-CoA, resulting in complete carbon conversion (3). Based on this, a concept called acetogenic mixotrophy was recently proposed to improve biofuel and biochemical yields (25). The acsA KO strain constructed in this study provides a unique opportunity to examine the effect of a disabled WLP on a microbe that normally performs acetogenic mixotrophy. The growth of the acsA KO strain on fructose was significantly impaired, highlighting the role of CODH/ACS in biomass formation during heterotrophic growth.
Acetate production, which generates an ATP per molecule of acetyl-CoA via substrate level phosphorylation, is one of the hallmarks of acetogens (3). Hence, it is surprising that the acsA KO strain with unperturbed phosphotransacetylase (pta) and acetate kinase (ack) genes only transiently produced trace amount of acetate while growing on fructose. Acetate is often viewed as an undesirable by-product during biofuel or commodity chemical production, so there are reports in the literature of attempts to engineer strains that produce no acetate. Most studies tried to block carbon flow by inactivating the key acetate-forming enzyme-encoding genes pta and/or ack (e.g., in Clostridium acetobutylicum [26], Clostridium tyrobutyricum [27], or E. coli [28]), but acetate production was not abolished and growth was reduced instead.
Instead of producing acetate and biomass, the acsA KO strain produced significantly more ethanol, 2,3-butanediol, and lactate than the WT while growing on fructose. Production of these products serves as an alternative sink to the disabled WLP (due to acsA inactivation) for reducing equivalents generated by glycolysis (Fig. 5), resulting in a redistribution of electron and carbon fluxes. By a similar approach, acetate production could be significantly reduced and carbon redistributed to ethanol when inactivating hydrogenase maturation proteins (and, therefore, hydrogenase activity) in Clostridium thermocellum (29). Certain acetogens, including C. autoethanogenum, harbor the enzyme aldehyde:ferredoxin oxidoreductase (14), which could reduce acetic acids into acetaldehyde using reduced ferredoxin, followed by the formation of ethanol via ethanol dehydrogenase. Accordingly, the lack of acetate production in the acsA KO strain could be due to an increase in the rate of acetic acid reduction. Collectively, our results demonstrated that although a functional WLP allows higher carbon utilization efficiency during heterotrophic growth by fixing CO2 into biomass and acetate, the biosynthesis of ethanol, 2,3-butanediol, and lactate is significantly reduced in the absence of an additional reductant (e.g., H2 or CO).
The acsA gene of C. autoethanogenum is uniquely truncated because of an internal TGA stop codon. The acsA genes of five other clostridial acetogens, Clostridium ljungdahlii (30), “Clostridium ragsdalei” (12), Clostridium carboxidivorans P7 (31), Clostridium aceticum (32), and Clostridium difficile 630 (33), with identical operon topologies showed no gene-splitting event (see Fig. S5A in the supplemental material). Instead, a TCA serine codon is present in these acetogens. Given the essential role of acsA in acetogenesis, it was of interest to determine whether posttranscriptional mechanisms allowed translational readthrough beyond the internal stop codon in C. autoethanogenum. One such mechanism is the incorporation of selenocysteine at the TGA codon, which is generally reliant on a characteristic bacterial selenocysteine insertion sequence (bSECIS) immediately downstream of the stop codon. A bSECIS element was not detected in the C. autoethanogenum acsA gene by using an algorithm (34), but such an element was uncovered via manual examination (see Fig. S5B in the supplemental material). The catalytic activities of many selenoproteins are often superior to those of their cysteine-dependent counterparts (35).
By investigating the translational pattern of FLAG-tagged AcsA protein by Western blot analysis, we showed that a partial and inefficient translational readthrough event occurs in C. autoethanogenum as formation of the truncated AcsA protein is the main product. It is not clear whether the ability of C. autoethanogenum to generate mature and truncated AcsA is a novel regulatory mechanism for an as-yet-unknown physiological purpose or whether it poses a handicap that hinders acetogenesis. A search of the curated genome (21) revealed 52 CDSs with TGA stop codons closely followed by an in-frame CDS (see Table S1 in supplemental material), including a selenocysteine formate dehydrogenase (36). Thus, acsA may not be the only C. autoethanogenum gene with an internally translated stop codon.
In contrast to C. autoethanogenum, plasmid expression of the FLAG-tagged AcsA protein in E. coli (a model organism for selenocysteine formation and incorporation) did not result in the formation of a translational readthrough product. This result indicates that the translational readthrough event in acsA is C. autoethanogenum specific and is unlikely to involve selenocysteine incorporation. Stop codon readthrough is not uncommon and especially prevalent for the UGA codon, while UAA is a more efficient translational stop signal (37). When the TAA stop codon of acsA replaced the internal TGA stop codon, translation of the full-length AcsA peptide in C. autoethanogenum and E. coli was completely eliminated. Stop codon readthrough depends on the competition between a release factor and a near-cognate tRNA (37). The genome of C. autoethanogenum encodes one tRNA with anticodon 5ʹ-CCA-3ʹ (CAETHG_R0046) for tryptophan.
In this study, inactivation of either cooS1 or cooS2 moderately lowered biomass formation during growth on CO but did not negatively impact growth on H2-CO2, highlighting functional redundancies in acetogens that harbor multiple CODH isogenes. The orthologous cooS1 and cooS2 genes of C. ljungdahlii were reported to be expressed at lower levels when the bacterium was grown autotrophically on CO rather than heterotrophically on fructose, leading the authors to hypothesize that monofunctional CODHs do not significantly contribute to the oxidation of CO, but it is the bifunctional CODH/ACS complex that is mainly responsible (16, 38). In agreement with this hypothesis, during mixotrophic growth on fructose plus CO, C. autoethanogenum WT, but not the acsA KO strain (which still carries the functional cooS1 and cooS2 genes), was able to oxidize CO.
During growth on H2-CO2, the cooS1 KO strain, surprisingly, grew without an apparent lag phase and reached twice the OD600 of the WT strain. Since the reduced [CO] moiety is enclosed within the CODH/ACS complex to lower internal thermodynamic barriers during acetyl-CoA synthesis (39–41), the presence of another CODH may act as a competitor for CO2. The inactivation of cooS1 would, therefore, increase the efficiency of the CODH/ACS enzyme complex, leading to the observed enhanced growth on H2-CO2, and may represent a metabolic engineering strategy to improve gas utilization efficiency in acetogens that harbor multiple CODH-encoding isogenes.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
For the bacterial strains used in this study, see Table S2 in the supplemental material. The E. coli strains used for general plasmid propagation, cloning, and conjugation were cultivated at 37°C in LB medium in the presence of antibiotics (25 µg/ml chloramphenicol, 100 µg/ml spectinomycin). C. autoethanogenum DSM 10061 was purchased from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany, and cultivated under strict anaerobic conditions in CaGM medium.
CaGM growth medium contains (per liter) 0.25 g of NH4Cl, 0.1 g of KCl, 0.2 g of KH2PO4, 0.2 g of MgSO4 ⋅ 7H2O, 0.02 g of CaCl2 ⋅ 2H2O, 1 g of yeast extract, 0.5 ml of 2-g/liter resazurin, 20 g of 2-(N-morpholino)ethanesulfonic acid (MES), 0.05 g of Fe(SO4)2 ⋅ 7H2O, 0.25 g of sodium acetate ⋅ 3H2O, 0.05 g of nitrilotriacetic acid (NTA), 10 g of fructose (only for heterotrophic growth), 10 ml of a trace element (TSE) solution, and 10 ml of Wolfe’s vitamin solution. The composition of the TSE solution (per liter) was 2 g of nitrilotriacetic acid, 1 g of MnSO4 ⋅ H2O, 0.8 g of Fe(SO4)2(NH4)2 ⋅ 6H2O, 0.2 g of CoCl2 ⋅ 6H2O, 0.2 mg of ZnSO4 ⋅ 7H2O, 0.02 g of CuCl2 ⋅ 2H2O, 0.02 g of NaMoO4 ⋅ 2H2O, 0.02 g of Na2SeO3, 0.02 g of NiCl2 ⋅ 6H2O, and 0.02 g of Na2WO4 ⋅ 2H2O. The vitamin solution composition (per liter) was 2 mg of biotin, 2 mg of folic acid, 10 mg of pyridoxine hydrochloride, 5 mg of thiamine HCl, 5 mg of riboflavin, 5 mg of nicotinic acid, 5 mg of calcium pantothenate, 0.1 mg of vitamin B12, 5 mg of p-aminobenzoic acid, and 5 mg of thioctic acid. The medium was prepared anaerobically, and the pH of the medium was adjusted to 5.8 before sterilization. Prior to inoculation, 100 ml of CaGM medium was reduced with 1 ml of reducing agent 1 (4 g of cysteine HCl/100 ml of water) and 1 ml of reducing agent 2 (7.64 g of NTA, 5.33 g of Na2CO3, and 8.5 ml of TiCl3/100 ml of water).
Cell growth on liquid medium was monitored spectrophotometrically by measuring optical density at 600 nm (OD600). Changes in headspace pressure were measured with Rugged Digital Pressure Gauge DPG120 (Omega Engineering). For growth of C. autoethanogenum on agar plates, YTF solid medium (10 g/liter fructose, 10 g/liter yeast extract, 16 g/liter tryptone, 0.2 g/liter sodium chloride, 15 g/liter bacteriological agar [Oxoid], pH 5.8), with antibiotics (7.5 µg/ml thiamphenicol, 6 µg/ml clarithromycin) where appropriate, was used. All mutagenesis work was performed inside an anaerobic workstation at 37°C (Don Whitley Scientific Ltd.). For strain comparisons, three or four biological replicates of WT or recombinant C. autoethanogenum strains were grown in 250-ml serum bottles containing 50 ml of CaGM medium with 10 g/liter fructose, 200 kPa CO, 10 g/liter fructose plus 200 kPa CO, or 130 kPa H2 plus 70 kPa CO2 as the growth substrate. Incubation at 37°C was done with agitation (225 rpm) inside New Brunswick Innova shakers (Eppendorf). A standardized 0.5-OD600 equivalent of exponentially growing cultures was used as the inoculum.
DNA manipulation.
DNA manipulation and cloning were carried out according to standard techniques described by Sambrook and Russell (42). Genomic DNA from C. autoethanogenum was isolated with a DNeasy Blood and Tissue kit (Qiagen) for PCR diagnostics. For Southern blot analysis, genomic DNA of C. autoethanogenum was extracted as described by Bertram and Dürre (43). Plasmid DNA from C. autoethanogenum was isolated with a QIAprep Spin Miniprep kit (Qiagen) with the supplementation of 20 mg/ml chicken lysozyme into lysis buffer and incubation at 37°C for 30 min before proceeding to downstream procedures. PCR was carried out with Phusion DNA polymerase (NEB) or Q5 DNA polymerase (NEB). For the primers used in this study, see Table S3 in the supplemental material. Primers were designed with Geneious (Biomatters) and synthesized by Sigma-Aldrich or Eurofins. Sanger sequencing of plasmids and amplicons was carried out by Source Bioscience (United Kingdom).
Plasmid vectors.
All of the plasmids used in this study were derived from the pMTL80000 series of modular E. coli-Clostridium shuttle vectors (44) (see Table S4 in the supplemental material). For the construction of plasmid pMTL83151-PacsA, the promoter region of C. autoethanogenum acsA (CAETHG_1621) was amplified with oligonucleotides PacsA-NotI-F and PacsA-NdeI-R and then cloned into plasmid pMTL83151 (44) by using the NotI and NdeI restriction sites. To construct the acsA overexpression/complementation plasmid, acsAfull (CAETHG1620-1621) was first subjected to SOE-PCR (45) with oligonucleotides listed in Table S3 in the supplemental material to remove an internal NdeI site at nucleotide position 342 of CAETHG_1620, resulting in a change in the nucleotide sequence from CATATG to CACATG while retaining the same encoded amino acid sequence. Following cleavage with NdeI and SacI, this amplicon was cloned into plasmid pMTL83151-PacsA to generate plasmid pMTL83151-PacsA-acsAfull.
A FLAG tag sequence (encoding the amino acid sequence DYKDDDDK) was fused to either the N or the C terminus of acsA, which was then cloned into plasmid pMTL83151 to generate four plasmid variants to examine the C. autoethanogenum acsA translation pattern. The first plasmid, pMTL83151-PacsA-FLAG-acsA(TGA), has an N-terminally FLAG-tagged acsA gene. It was constructed by initial PCR amplification of native acsA with oligonucleotides NcoI-FLAG-acsA-F and acsA-HindIII-R and cloning of the fragment generated into plasmid pMTL83151 by using the NcoI and HindIII restriction sites. This was followed by PCR amplification of a DNA fragment encompassing the native PacsA promoter with primers PacsA-SacI-F and PacsA-NcoI-R and its cloning between the SacI and NcoI sites of plasmid pMTL83151. The second plasmid, pMTL83151-PacsA-acsA(TGA)-FLAG, was generated by the amplification of acsA and its native promoter with oligonucleotides PacsA-SacI-F and acsA-FLAG-BamHI-R and cloning of the product into plasmid pMTL83151 by using the SacI and BamHI restriction sites.
The third plasmid, pMTL83151-PacsA-acsA(TCA)-FLAG, has the internal TGA stop codon of C. autoethanogenum acsA mutated to a TCA serine codon. To assemble this plasmid, SOE-PCR was performed with oligonucleotides PacsA-SacI-F, acsA(TCA)-SOE-B, acsA(TCA)-SOE-C, and acsA-FLAG-BamHI-R and then the product was cloned into plasmid pMTL83151 by using the SacI and BamHI restriction sites. Similarly, the fourth plasmid, pMTL83151-PacsA-acsA(TAA)-FLAG, consists of an acsA variant that has the internal TGA codon mutated to another stop codon, TAA. This plasmid was constructed by first performing SOE-PCR with primers PacsA-SacI-F, acsA(TAA)-SOE-B, acsA(TAA)-SOE-C, and acsA-FLAG-BamHI-R and then cloning the product into plasmid pMTL83151 by using the SacI and BamHI restriction sites. The cloned promoter and CDS insert in all of the above-described plasmids were verified by Sanger sequencing.
For the construction of ClosTron retargeting plasmids, the appropriate intron-targeting regions within cooS1, cooS2, and acsA were generated in silico as previously described (18) using a web-based Perutka algorithm (46). DNA 2.0, Inc., then synthesized the 344-bp intron-targeting region and cloned it into ClosTron vector pMTL007C-E2 (18) by using restriction sites HindIII and BsrGI, resulting in plasmids pMTL007C-E2::cooS1_601s, pMTL007C-E2::cooS2_529s, and pMTL007C-E2::acsA_143s (see Table S4 in the supplemental material).
Plasmid transfer into C. autoethanogenum.
Plasmids were transformed into E. coli donor strain CA434 and then conjugated into C. autoethanogenum by previously established methods (15, 47, 48). Thiamphenicol (7.5 µg/ml) was used to select for catP-based plasmids. Trimethoprim (10 µg/ml) was used to counterselect against E. coli CA434 after conjugation. For the validation of plasmid overexpression and plasmid complementation strains, plasmids were isolated from C. autoethanogenum transconjugants and subsequently transformed into E. coli cells before restriction digestion analysis of the rescued plasmids was carried out (see Fig. S6 in supplemental material). The 16S rRNA gene was also amplified from the genomic DNA of transconjugants with oligonucleotides univ-0027-F and univ-1492-R and then Sanger sequenced for verification.
Construction of C. autoethanogenum ClosTron strains.
Following conjugation of ClosTron retargeting plasmids into C. autoethanogenum by using E. coli strain CA434 as the donor, thiamphenicol- and trimethoprim-resistant colonies were transferred onto solid YTF medium supplemented with 6 µg/ml clarithromycin to select for intron insertions at target loci and repeatedly streaked onto the same selective medium until plasmid loss was demonstrated, as evident in loss of the ability to grow on medium supplemented with thiamphenicol. Genomic DNA was extracted from the clarithromycin-resistant colonies and subjected to PCR screening with locus-specific flanking primers (see Table S3 in the supplemental material) to identify clones that produced an amplicon 1.8 kb larger than that of the WT control (indicative of ClosTron insertion at specified DNA locus). Sanger sequencing of the ClosTron amplicons was performed to validate the location of ClosTron insertion. As final verification, Southern blot analysis was performed with a digoxigenin High-Prime DNA labeling and detection kit (Roche) as instructed by the manufacturer to ensure that only one ClosTron insertion had occurred in each KO strain. Clones with multiple ClosTron insertions were omitted from downstream studies. For the complementation of the acsA KO strain, plasmid pMTL83151-PacsA-acsAfull was conjugated into this strain and verified by restriction digestion analysis of rescued plasmids from the transconjugants (data not shown).
Preparation of crude lysates.
Transformed E. coli strain XL1-Blue MRFʹ and C. autoethanogenum were cultured in 50-ml Falcon tubes inside an anaerobic cabinet. Transformed E. coli was cultivated in 50 ml of LB medium supplemented with 20 mM glucose, 10 µM Na2SeO3, and 25 µg/ml chloramphenicol for 23 to 28 h. C. autoethanogenum transconjugants were cultivated in 10 ml of YTF medium supplemented with 15 µg/ml thiamphenicol for 44 to 51 h.
Cell pellets were harvested by centrifugation at 4°C at 7,197 × g for 10 min and then resuspended in 300 µl of lysis buffer (50 mM Tris-HCl [pH 7.4], 100 mM NaCl) containing fresh 20 mg/ml chicken lysozyme. Following incubation at 37°C for 45 min, the cell suspensions were sonicated with a Bioruptor Plus (Diagenode) for 60 cycles of 30 s of sonication and 30 s of rest per cycle at 4°C. Following ultracentrifugation at 20,238 × g for 5 min at 4°C, the supernatant was harvested as the soluble fraction, whereas the pellet represented the insoluble fraction and was resuspended in 300 µl of lysis buffer without chicken lysozyme. Protein contents were quantified with Bradford reagent (Sigma-Aldrich) with bovine serum albumin as the standard. Both the soluble and insoluble fractions of crude lysates were stored at −20°C for further analysis.
Western blot analysis.
Cell lysates and purified proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). NuPAGE LDS sample buffer (Invitrogen) and 83.3 mM (final concentration) dithiothreitol were added to each sample, and it was boiled at 100°C for 5 min to denature the proteins. The samples and Precision Plus Protein Kaleidoscope Standards (Bio-Rad Laboratories) were then loaded onto 4 to 12% NuPAGE Bis-Tris gels (Invitrogen) in the XCell SureLock Mini-Cell Electrophoresis System (Life Technologies), and NuPAGE MES running buffer (Invitrogen) was added to cover the electrodes. The samples were then subjected to electrophoresis at 150 V for 140 min to separate the proteins, after which the gel was removed from the cast, laid onto Amersham Hybond ECL nitrocellulose membrane (GE Healthcare), and then fitted into an XCell II Blot Module (Life Technologies). To transfer proteins to the membrane, Novex transfer buffer (10% [vol/vol] methanol) was added to the blot module and subjected to electrophoresis at 30 V for 2 h.
Following disassembly of the blot module, the membrane was blocked in 30 ml of TBS buffer (50 mM Tris-HCl, 150 mM NaCl [pH 7.5], 3% [wt/vol] skim milk) at room temperature for 1 h with mild agitation on a shaker. After removal of the used TBS buffer, the membrane (covered in foil to protect it from light) was subjected to overnight incubation in 30 ml of TBS buffer containing 10 µl of anti-FLAG M2-Peroxidase (horseradish peroxidase) monoclonal antibody (Sigma-Aldrich) at room temperature with mild agitation. Following three 5-min washes in TBST (50 mM Tris-HCl, 150 mM NaCl, 0.1% [vol/vol] Tween [pH 7.5]), 4 ml of 3,3′,5,5′-tetramethylbenzidine detection substrate (Sigma-Aldrich) was added to the membrane, which was incubated at room temperature for 5 min before gentle rinsing with deionized H2O. The membrane was air dried for 30 min before an image was captured with an EOS 600D DSLR camera (Canon).
Analytical chemistry.
Analysis of metabolites was performed with the Varian ProStar HPLC (high-performance liquid chromatography) system equipped with a refractive index detector operated at 30°C and an Aminex HPX-87H column (1,300 by 7.8 mm; particle size, 9 µm; Bio-Rad Laboratories) kept at 30°C. Slightly acidified water was used (0.005 M H2SO4) as the mobile phase with a flow rate of 0.5 ml/min. To remove proteins and other cell residues, samples were centrifuged at 20,238 × g for 5 min and the supernatant was filtered with Spartan 13/0.2 RC filters. Ten microliters of the supernatant was then injected into the HPLC system for analysis. Measurements of headspace gas composition were carried out on a Varian CP-4900 micro gas chromatograph with two installed channels. Channel 1 was a 10-m Mol-sieve column running at 70°C with 200 kPa argon and a backflush time of 4.2 s, while channel 2 was a 10-m PPQ column running at 90°C with 150 kPa helium and no backflush. The injector temperature for both channels was 70°C. The run time was set to 120 s, but all of the peaks of interest eluted before 100 s.
Alignment of acsA nucleotide sequences.
The nucleotide sequences of acsA from C. autoethanogenum (GenBank accession no. NC_022592), C. ljungdahlii (CP001666), “C. ragsdalei” (HQ876032), C. carboxidivorans P7 (HM590563), C. aceticum (CP009687), and C. difficile 630 (NC_009089) were obtained from NCBI. Multiple global sequence alignments with free-end gaps were performed with Geneious (Biomatters) version 6.1.7.
Data analysis and presentations.
Statistical analysis was performed and graphically presented results were prepared with GraphPad Prism. Two-tailed, unpaired, parametric Student t tests were employed for comparisons of means.
SUPPLEMENTAL MATERIAL
Screening and validation of cooS1, cooS2, and acsA KO strains. (A) Gel electrophoresis of products of PCRs with exon-spanning primers. Lanes: M, NEB 2-log DNA ladder; 2 to 6, cooS1 KO clones; 9 to 11, cooS2 KO clones; 14 to 16, acsA KO clones; 1, 8, and 13, nontemplate controls for cooS1, cooS2, and acsA exon-spanning primer pairs, respectively; 7, 12, and 17, WT controls for cooS1, cooS2, and acsA exon-spanning primer pairs, respectively. (B) Southern blot analysis of HindIII-digested genomic DNA of cooS1 KO clones (lanes 18 to 21), cooS2 KO clones (lanes 22 to 26), and acsA KO clones (lanes 27 to 30). Download
Growth and metabolite profiles of the complemented acsA KO strain and plasmid control strains on 200 kPa CO. Panels: A, growth profile; B, acetate profile, C, ethanol profile, D, 2,3-butanediol profile. Black circles, plasmid control (n = 4); red squares, acsA KO strain (n = 3); blue triangles, complemented strain (n = 3). Error bars show the standard error of the mean. Download
Effect of acsAfull overexpression on growth and metabolite profiles of C. autoethanogenum on 200 kPa CO. Panels: A, growth profile, B, acetate profile, C, ethanol profile, D, 2,3-butanediol profile. Orange triangles, acsAfull overexpression strain (n = 3); black circles, plasmid control strain (n = 4). Error bars show the standard error of the mean. Download
Effect of acsAfull overexpression on growth and metabolite profiles of C. autoethanogenum on 10 g/liter fructose. Panels: A, growth profile, B, acetate profile, C, ethanol profile. Orange triangles, acsAfull overexpression strain; black circles, plasmid control strain. n = 3. Error bars show the standard error of the mean. Download
Nucleotide sequence alignment of acsA and internal stop codon in C. autoethanogenum. (A) Nucleotide sequence alignment of the acsA CDSs (GenBank accession number) of C. autoethanogenum (C_auto; NC_022592), C. ljungdahlii (C_ljung; CP001666), C. ragsdalei (C_rags; HQ876032), C. carboxidivorans P7 (C_carb; HM590563), C. difficile 630 (C_diff; NC_009089), and C. aceticum (C_acetic; CP009687). Variations in the nucleotide sequence of C. autoethanogenum are marked in black or as gaps in the gray sequence bar above the red annotations. (B) Inset of panel A highlighting internal the TGA stop codon and putative bacterial selenocysteine insertion sequence (bSECIS) in acsA of C. autoethanogenum. The image was generated with Geneious 6.1.7 (Biomatters Ltd.). Download
Restriction digestion (SacI and BamHI) analysis of rescued acsA plasmids from C. autoethanogenum transconjugants. Lanes: M, NEB 2-log DNA ladder; 1 to 4, pMTL83151-PacsA-FLAG-acsA(TGA) clones; 5 to 12, pMTL83151-PacsA-acsA(TGA)-FLAG clones; 13 to 20, pMTL83151-PacsA-FLAG-acsA(TCA) clones; 21 to 24, pMTL83151-PacsA-FLAG-acsA(TAA) clones. Download
C. autoethanogenum CDSs (CAETHG_) with a TGA stop codon that is immediately followed by an in-frame gene.
Bacterial strains used in this study.
Oligonucleotides used in this study.
Plasmids used in this study.
ACKNOWLEDGMENTS
We thank the following investors in LanzaTech’s technology: Stephen Tindall; Khosla Ventures; Qiming Venture Partners; Softbank China; the Malaysian Life Sciences Capital Fund; Mitsui; Primetals; CICC Growth Capital Fund I, L.P.; and the New Zealand Superannuation Fund. N.P.M., K.W., and A.M. acknowledge the financial support of the United Kingdom Biotechnology and Biological Sciences Research Council (grant BB/K00283X/1). LanzaTech has a commercial interest in gas fermentation with C. autoethanogenum. We have no competing financial interests to declare.
N.P.M., S.D.S., K.W., and F.L. conceived the project. F.L., K.W., A.M.H., and M.K. designed the experiments. F.L. performed the experiments with assistance from A.M.H. F.L., K.W., M.K., A.M.H., and N.P.M. analyzed the results. F.L., M.K., A.M.H., and N.P.M. wrote the manuscript. All of us discussed the results and commented on the manuscript.
Footnotes
Citation Liew F, Henstra AM, Winzer K, Köpke M, Simpson SD, Minton NP. 2016. Insights into CO2 fixation pathway of Clostridium autoethanogenum by targeted mutagenesis. mBio 7(3):e00427-16. doi:10.1128/mBio.00427-16.
REFERENCES
- 1.Fast AG, Papoutsakis ET. 2012. Stoichiometric and energetic analyses of non-photosynthetic CO2-fixation pathways to support synthetic biology strategies for production of fuels and chemicals. Curr Opin Chem Eng 1:380–395. doi: 10.1016/j.coche.2012.07.005. [DOI] [Google Scholar]
- 2.Martin WF. 2012. Hydrogen, metals, bifurcating electrons, and proton gradients: the early evolution of biological energy conservation. FEBS Lett 586:485–493. doi: 10.1016/j.febslet.2011.09.031. [DOI] [PubMed] [Google Scholar]
- 3.Drake HL, Küsel K, Matthies C. 2006. Acetogenic prokaryotes, p 354–420. In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (ed), The prokaryotes, 3rd ed. Springer Verlag, New York, NY. [Google Scholar]
- 4.Liew FM, Köpke M, Simpson SD. 2013. Gas fermentation for commercial biofuels production. In Fang Z (ed), Liquid, gaseous and solid biofuels—conversion techniques. InTech; Rijeka, Croatia. [Google Scholar]
- 5.Handler RM, Shonnard DR, Griffing EM, Lai A, Palou-Rivera I. 2016. Life cycle assessments of ethanol production via gas fermentation: anticipated greenhouse gas emissions for cellulosic and waste gas feedstocks. Ind Eng Chem Res 55:3253–3261. doi: 10.1021/acs.iecr.5b03215. [DOI] [Google Scholar]
- 6.McGlade C, Ekins P. 2015. The geographical distribution of fossil fuels unused when limiting global warming to 2 C. Nature 517:187–190. doi: 10.1038/nature14016. [DOI] [PubMed] [Google Scholar]
- 7.Grahame DA, Demoll E. 1995. Substrate and accessory protein requirements and thermodynamics of acetyl-CoA synthesis and cleavage in Methanosarcina barkeri. Biochemistry 34:4617–4624. doi: 10.1021/bi00014a015. [DOI] [PubMed] [Google Scholar]
- 8.Oelgeschläger E, Rother M. 2008. Carbon monoxide-dependent energy metabolism in anaerobic bacteria and archaea. Arch Microbiol 190:257–269. doi: 10.1007/s00203-008-0382-6. [DOI] [PubMed] [Google Scholar]
- 9.Darnault C, Volbeda A, Kim EJ, Legrand P, Vernède X, Lindahl PA, Fontecilla-Camps JC. 2003. Ni-Zn-[Fe4-S4] and Ni-Ni-[Fe4-S4] clusters in closed and open subunits of acetyl-CoA synthase/carbon monoxide dehydrogenase. Nat Struct Biol 10:271–279. doi: 10.1038/nsb912. [DOI] [PubMed] [Google Scholar]
- 10.Doukov TI, Iverson TM, Seravalli J, Ragsdale SW, Drennan CL. 2002. A Ni-Fe-Cu center in a bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase. Science 298:567–572. doi: 10.1126/science.1075843. [DOI] [PubMed] [Google Scholar]
- 11.Can M, Armstrong FA, Ragsdale SW. 2014. Structure, function, and mechanism of the nickel metalloenzymes, CO dehydrogenase, and acetyl-CoA synthase. Chem Rev 114:4149–4174. doi: 10.1021/cr400461p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Köpke M, Mihalcea C, Liew F, Tizard JH, Ali MS, Conolly JJ, Al-Sinawi B, Simpson SD. 2011. 2,3-Butanediol production by acetogenic bacteria, an alternative route to chemical synthesis, using industrial waste gas. Appl Environ Microbiol 77:5467–5475. doi: 10.1128/AEM.00355-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abrini J, Naveau H, Nyns E. 1994. Clostridium autoethanogenum, sp. nov., an anaerobic bacterium that produces ethanol from carbon monoxide. Arch Microbiol 161:345–351. doi: 10.1007/BF00303591. [DOI] [Google Scholar]
- 14.Brown SD, Nagaraju S, Utturkar S, De Tissera S, Segovia S, Mitchell W, Land ML, Dassanayake A, Köpke M. 2014. Comparison of single-molecule sequencing and hybrid approaches for finishing the genome of Clostridium autoethanogenum and analysis of CRISPR systems in industrial relevant clostridia. Biotechnol Biofuels 7:40. doi: 10.1186/1754-6834-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mock J, Zheng Y, Mueller AP, Ly S, Tran L, Segovia S, Nagaraju S, Köpke M, Dürre P, Thauer RK. 2015. Energy conservation associated with ethanol formation from H2 and CO2 in Clostridium autoethanogenum involving electron bifurcation. J Bacteriol 197:2965–2980. doi: 10.1128/JB.00399-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marcellin E, Behrendorff JB, Nagaraju S, DeTissera S, Segovia S, Palfreyman RW, Daniell J, Licona-Cassani C, Quek L, Speight R, Hodson MP, Simpson SD, Mitchell WP, Köpke M, Nielsen LK 6 January 2016. Low carbon fuels and commodity chemicals from waste gases—systematic approach to understand energy metabolism in a model acetogen. Green Chem doi: 10.1039/C5GC02708J. [DOI] [Google Scholar]
- 17.Heap JT, Pennington OJ, Cartman ST, Carter GP, Minton NP. 2007. The ClosTron: a universal gene knockout system for the genus Clostridium. J Microbiol Methods 70:452–464. doi: 10.1016/j.mimet.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 18.Heap JT, Kuehne SA Ehsaan M, Cartman ST, Cooksley CM, Scott JC, Minton NP. 2010. The ClosTron: mutagenesis in Clostridium refined and streamlined. J Microbiol Methods 80:49–55. doi: 10.1016/j.mimet.2009.10.018. [DOI] [PubMed] [Google Scholar]
- 19.Kuehne SA, Minton NP. 2012. ClosTron-mediated engineering of Clostridium. Bioengineered 3:247–254. doi: 10.4161/bioe.21004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Utturkar SM, Klingeman DM, Bruno-Barcena JM, Chinn MS, Grunden AM, Köpke M, Brown SD. 2015. Sequence data for Clostridium autoethanogenum using three generations of sequencing technologies. Sci Data 2:150014. doi: 10.1038/sdata.2015.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Humphreys CM, McLean S, Schatschneider S, Millat T, Henstra AM, Annan FJ, Breitkopf R, Pander B, Piatek P, Rowe P, Wichlacz AT, Woods C, Norman R, Blom J, Goesman A, Hodgman C, Barrett D, Thomas NR, Winzer K, Minton NP. 2015. Whole genome sequence and manual annotation of Clostridium autoethanogenum, an industrially relevant bacterium. BMC Genomics 16:1085. doi: 10.1186/s12864-015-2287-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allmang C, Krol A. 2006. Selenoprotein synthesis: UGA does not end the story. Biochimie 88:1561–1571. doi: 10.1016/j.biochi.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 23.Zinoni F, Birkmann A, Stadtman TC, Böck A. 1986. Nucleotide sequence and expression of the selenocysteine-containing polypeptide of formate dehydrogenase (formate-hydrogen-lyase-linked) from Escherichia coli. Proc Natl Acad Sci U S A 83:4650–4654. doi: 10.1073/pnas.83.13.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matschiavelli N, Oelgeschläger E, Cocchiararo B, Finke J, Rother M. 2012. Function and regulation of isoforms of carbon monoxide dehydrogenase/acetyl coenzyme A synthase in Methanosarcina acetivorans. J Bacteriol 194:5377–5387. doi: 10.1128/JB.00881-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fast AG, Schmidt ED, Jones SW, Tracy BP. 2015. Acetogenic mixotrophy: novel options for yield improvement in biofuels and biochemicals production. Curr Opin Biotechnol 33:60–72. doi: 10.1016/j.copbio.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 26.Kuit W, Minton NP, López-Contreras AM, Eggink G. 2012. Disruption of the acetate kinase (ack) gene of Clostridium acetobutylicum results in delayed acetate production. Appl Microbiol Biotechnol 94:729–741. doi: 10.1007/s00253-011-3848-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu X, Zhu Y, Yang ST. 2006. Construction and characterization of ack deleted mutant of Clostridium tyrobutyricum for enhanced butyric acid and hydrogen production. Biotechnol Prog 22:1265–1275. doi: 10.1021/bp060082g. [DOI] [PubMed] [Google Scholar]
- 28.Chang DE, Shin S, Rhee JS, Pan JG. 1999. Acetate metabolism in a pta mutant of Escherichia coli W3110: importance of maintaining acetyl coenzyme A flux for growth and survival. J Bacteriol 181:6656–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biswas R, Zheng T, Olson DG, Lynd LR, Guss AM. 2015. Elimination of hydrogenase active site assembly blocks H2 production and increases ethanol yield in Clostridium thermocellum. Biotechnol Biofuels 8:20. doi: 10.1186/s13068-015-0204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Köpke M, Held C, Hujer S, Liesegang H, Wiezer A, Wollherr A, Ehrenreich A, Liebl W, Gottschalk G, Dürre P. 2010. Clostridium ljungdahlii represents a microbial production platform based on syngas. Proc Natl Acad Sci U S A 107:13087–13092. doi: 10.1073/pnas.1004716107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bruant G, Lévesque M-J, Peter C, Guiot SR, Masson L. 2010. Genomic analysis of carbon monoxide utilization and butanol production by Clostridium carboxidivorans strain P7. PLoS One 5:e13033. doi: 10.1371/journal.pone.0013033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poehlein A, Cebulla M, Ilg MM, Bengelsdorf FR, Schiel-Bengelsdorf B, Whited G, Andreesen JR, Gottschalk G, Daniel R, Dürre P. 2015. The complete genome sequence of Clostridium aceticum: a missing link between Rnf- and cytochrome-containing autotrophic acetogens. mBio 6:e01168-15. doi: 10.1128/mBio.01168-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Köpke M, Straub M, Dürre P. 2013. Clostridium difficile is an autotrophic bacterial pathogen. PLoS One 8:e62157. doi: 10.1371/journal.pone.0062157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Gladyshev VN. 2005. An algorithm for identification of bacterial selenocysteine insertion sequence elements and selenoprotein genes. Bioinformatics 21:2580–2589. doi: 10.1093/bioinformatics/bti400. [DOI] [PubMed] [Google Scholar]
- 35.Johansson L, Gafvelin G, Arnér ES. 2005. Selenocysteine in proteins—properties and biotechnological use. Biochim Biophys Acta 1726:1–13. doi: 10.1016/j.bbagen.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 36.Wang S, Huang H, Kahnt J, Mueller AP, Köpke M, Thauer RK. 2013. NADP-specific electron-bifurcating [FeFe]-hydrogenase in a functional complex with formate dehydrogenase in Clostridium autoethanogenum grown on CO. J Bacteriol 195:4373–4386. doi: 10.1128/JB.00678-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tate WP, Mansell JB, Mannering SA, Irvine JH, Major LL, Wilson DN. 1999. UGA: a dual signal for “stop” and for recoding in protein synthesis. Biochemistry (Mosc.) 64:1342–1353. [PubMed] [Google Scholar]
- 38.Tan Y, Liu J, Chen X, Zheng H, Li F. 2013. RNA-seq-based comparative transcriptome analysis of the syngas-utilizing bacterium Clostridium ljungdahlii DSM 13528 grown autotrophically and heterotrophically. Mol Biosyst 9:2775–2784. doi: 10.1039/c3mb70232d. [DOI] [PubMed] [Google Scholar]
- 39.Maynard EL, Lindahl PA. 1999. Evidence of a molecular tunnel connecting the active sites for CO2 reduction and acetyl-CoA synthesis in acetyl-CoA synthase from Clostridium thermoaceticum. J Am Chem Soc 121:9221–9222. doi: 10.1021/ja992120g. [DOI] [Google Scholar]
- 40.Seravalli J, Ragsdale SW. 2000. Channeling of carbon monoxide during anaerobic carbon dioxide fixation. Biochemistry 39:1274–1277. doi: 10.1021/bi991812e. [DOI] [PubMed] [Google Scholar]
- 41.Bar-Even A. 2013. Does acetogenesis really require especially low reduction potential? Biochim Biophys Acta 1827:395–400. doi: 10.1016/j.bbabio.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, New York, NY. [Google Scholar]
- 43.Bertram J, Dürre P. 1989. Conjugal transfer and expression of streptococcal transposons in Clostridium acetobutylicum. Arch Microbiol 151:551–557. doi: 10.1007/BF00454874. [DOI] [Google Scholar]
- 44.Heap JT, Pennington OJ, Cartman ST, Minton NP. 2009. A modular system for Clostridium shuttle plasmids. J Microbiol Methods 78:79–85. doi: 10.1016/j.mimet.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 45.Warrens AN, Jones MD, Lechler RI. 1997. Splicing by overlap extension by PCR using asymmetric amplification: an improved technique for the generation of hybrid proteins of immunological interest. Gene 186:29–35. doi: 10.1016/S0378-1119(96)00674-9. [DOI] [PubMed] [Google Scholar]
- 46.Perutka J, Wang W, Goerlitz D, Lambowitz AM. 2004. Use of computer-designed group II introns to disrupt Escherichia coli DExH/D-box protein and DNA helicase genes. J Mol Biol 336:421–439. doi: 10.1016/j.jmb.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 47.Purdy D, O’Keeffe TA, Elmore M, Herbert M, McLeod A, Bokori-Brown M, Ostrowski A, Minton NP. 2002. Conjugative transfer of clostridial shuttle vectors from Escherichia coli to Clostridium difficile through circumvention of the restriction barrier. Mol Microbiol 46:439–452. doi: 10.1046/j.1365-2958.2002.03134.x. [DOI] [PubMed] [Google Scholar]
- 48.Williams DR, Young DI, Young M. 1990. Conjugative plasmid transfer from Escherichia coli to Clostridium acetobutylicum. J Gen Microbiol 136:819–826. doi: 10.1099/00221287-136-5-819. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Screening and validation of cooS1, cooS2, and acsA KO strains. (A) Gel electrophoresis of products of PCRs with exon-spanning primers. Lanes: M, NEB 2-log DNA ladder; 2 to 6, cooS1 KO clones; 9 to 11, cooS2 KO clones; 14 to 16, acsA KO clones; 1, 8, and 13, nontemplate controls for cooS1, cooS2, and acsA exon-spanning primer pairs, respectively; 7, 12, and 17, WT controls for cooS1, cooS2, and acsA exon-spanning primer pairs, respectively. (B) Southern blot analysis of HindIII-digested genomic DNA of cooS1 KO clones (lanes 18 to 21), cooS2 KO clones (lanes 22 to 26), and acsA KO clones (lanes 27 to 30). Download
Growth and metabolite profiles of the complemented acsA KO strain and plasmid control strains on 200 kPa CO. Panels: A, growth profile; B, acetate profile, C, ethanol profile, D, 2,3-butanediol profile. Black circles, plasmid control (n = 4); red squares, acsA KO strain (n = 3); blue triangles, complemented strain (n = 3). Error bars show the standard error of the mean. Download
Effect of acsAfull overexpression on growth and metabolite profiles of C. autoethanogenum on 200 kPa CO. Panels: A, growth profile, B, acetate profile, C, ethanol profile, D, 2,3-butanediol profile. Orange triangles, acsAfull overexpression strain (n = 3); black circles, plasmid control strain (n = 4). Error bars show the standard error of the mean. Download
Effect of acsAfull overexpression on growth and metabolite profiles of C. autoethanogenum on 10 g/liter fructose. Panels: A, growth profile, B, acetate profile, C, ethanol profile. Orange triangles, acsAfull overexpression strain; black circles, plasmid control strain. n = 3. Error bars show the standard error of the mean. Download
Nucleotide sequence alignment of acsA and internal stop codon in C. autoethanogenum. (A) Nucleotide sequence alignment of the acsA CDSs (GenBank accession number) of C. autoethanogenum (C_auto; NC_022592), C. ljungdahlii (C_ljung; CP001666), C. ragsdalei (C_rags; HQ876032), C. carboxidivorans P7 (C_carb; HM590563), C. difficile 630 (C_diff; NC_009089), and C. aceticum (C_acetic; CP009687). Variations in the nucleotide sequence of C. autoethanogenum are marked in black or as gaps in the gray sequence bar above the red annotations. (B) Inset of panel A highlighting internal the TGA stop codon and putative bacterial selenocysteine insertion sequence (bSECIS) in acsA of C. autoethanogenum. The image was generated with Geneious 6.1.7 (Biomatters Ltd.). Download
Restriction digestion (SacI and BamHI) analysis of rescued acsA plasmids from C. autoethanogenum transconjugants. Lanes: M, NEB 2-log DNA ladder; 1 to 4, pMTL83151-PacsA-FLAG-acsA(TGA) clones; 5 to 12, pMTL83151-PacsA-acsA(TGA)-FLAG clones; 13 to 20, pMTL83151-PacsA-FLAG-acsA(TCA) clones; 21 to 24, pMTL83151-PacsA-FLAG-acsA(TAA) clones. Download
C. autoethanogenum CDSs (CAETHG_) with a TGA stop codon that is immediately followed by an in-frame gene.
Bacterial strains used in this study.
Oligonucleotides used in this study.
Plasmids used in this study.