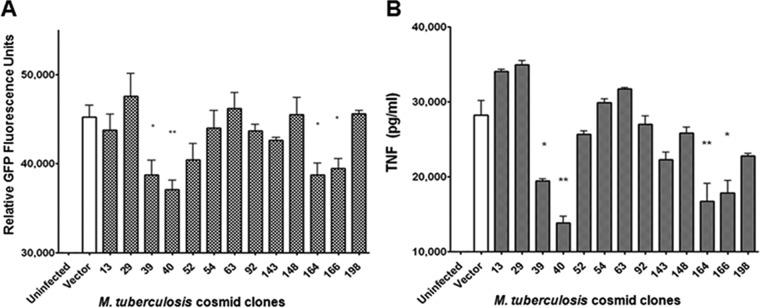
FIG 2 .
Screening of an M. tuberculosis cosmid library generated in heterologous host M. smegmatis strain mc2155 yielded M. tuberculosis cosmid clones with a TNF-downregulating phenotype. Clone C10 macrophages were infected at a multiplicity of infection (MOI) of 10 with M. smegmatis containing either empty vector (pYUB412 containing no H37Rv DNA) or M. tuberculosis cosmid clones (a total of 105 clones covering approximately half of the M. tuberculosis genome were screened). From the original transformation reaction, three independent colonies were picked for each cosmid clone. Therefore, three sets of the 105 cosmid clone library (named “a,” “b,” and “c”) were stocked. The screen of the 105 cosmid clones (from set “a”) was conducted in triplicate in 96-well plates, and the experiments were repeated 2 to 3 times. Clones with the TNF-downregulating phenotype underwent a second round of C10 screening with a similar setup. Clones that consistently induced GFP signal at a level lower than that generated by M. smegmatis transformed with empty vector were identified. To stringently test the TNF-downregulating phenotype of these second-round hits, corresponding independent clones from set “b” were tested. This third round of screening identified four clones (clones 39, 40, 164, and 166) with a consistent TNF-downregulating phenotype as assessed by GFP fluorescence signal (A) or ELISA measurement of the level of TNF in the supernatants of infected C10 cultures (B). There was a 100% concordance between the C10 fluorescence signal readout and the ELISA-based quantification of TNF production. Values are the means of the results of triplicates ± standard deviations (SD) and are representative of three separate experiments. *, P < 0.05; **, P < 0.005.