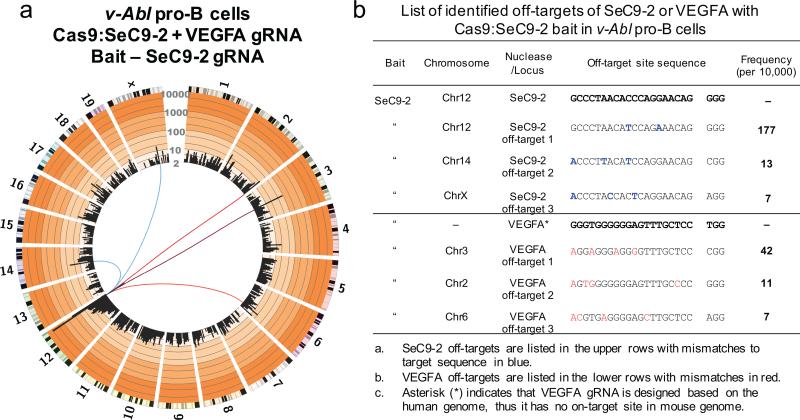
Figure 5. Universal bait detection of off-targets for designed VEGFA gRNA.
(a) Circos plot (circos.ca) on custom log scale showing genome-wide profile of Cas9:SeC9-2 junctions in cycling v-Abl pro-B cells. Bin size is 5 Mb and 8751 unique junctions are shown from 2 independent libraries. Chromosomes are displayed as centromere to telomere in a clock-wise orientation. Blue lines link SeC9-2 off-targets to bait break-site while red lines link VEGFA off-targets to bait break-site. (b) List of identified off-targets for SeC9-2 or VEGFA. The off-targets were identified by MACS2 as described previously6. v-Abl cells (3×106) were nucleofected with a combined total of 3 μg plasmid DNA in SF solution using the DN-100 program (Lonza) and collected 48 hours post nucleofection.