Fig. 5.
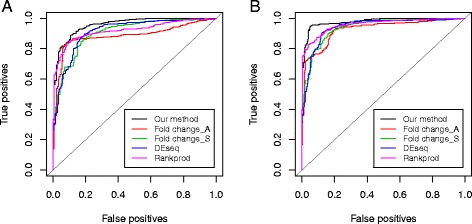
Comparison of discriminative power in the analysis of synthetic microRNA data. Figures show the percentage of correct and incorrect calls at various thresholds for two levels of classification stringency. The AUCs are shown in Additional file 1: Table S3. a. Genes with no fold change as true non-DEGs and the rest as true DEGs. b. Genes with a log2 fold change less than ±0.5 as true non-DEGs and the rest as true DEGs. Fold changes measured by microarray and RNA-seq are denoted as Fold change_A and Fold change_S, respectively
