Figure 1.
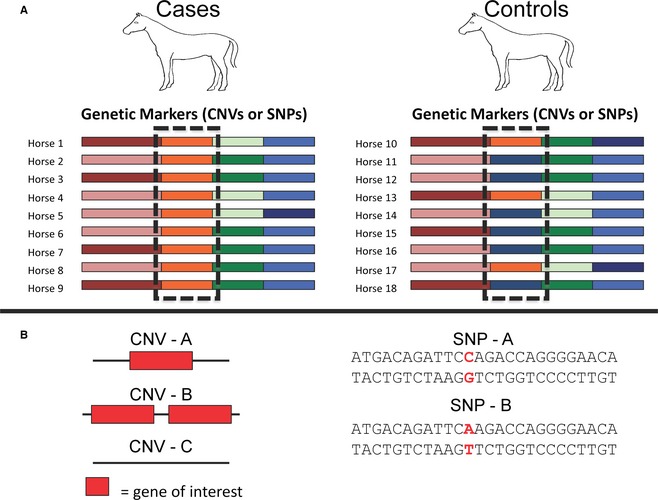
Association studies, CNVs, and SNPs. (A) The colored blocks indicate different alleles or haplotypes present in the horse genome. These have been identified by either a CNV or a SNP but any type of genetic variation can be used to identify alleles. The boxed regions show a greater frequency of the orange allele in the cases compared to the controls. The increased frequency of this allele in the cases suggests that it may harbor a variant(s) causing or contributing to the associated phenotype. (B) CNV – A represents a single copy of a gene; CNV – B represents a duplication of the gene; and, CNV – C represents a deletion of the gene. These examples demonstrate how CNVs can affect a single gene and can be used to identify different alleles in a population. SNPs, represented as red bases, offer the ability to identify alleles because of their polymorphic nature. Either type of genetic variation can be used in a GWAS to identify disease‐associated alleles.
