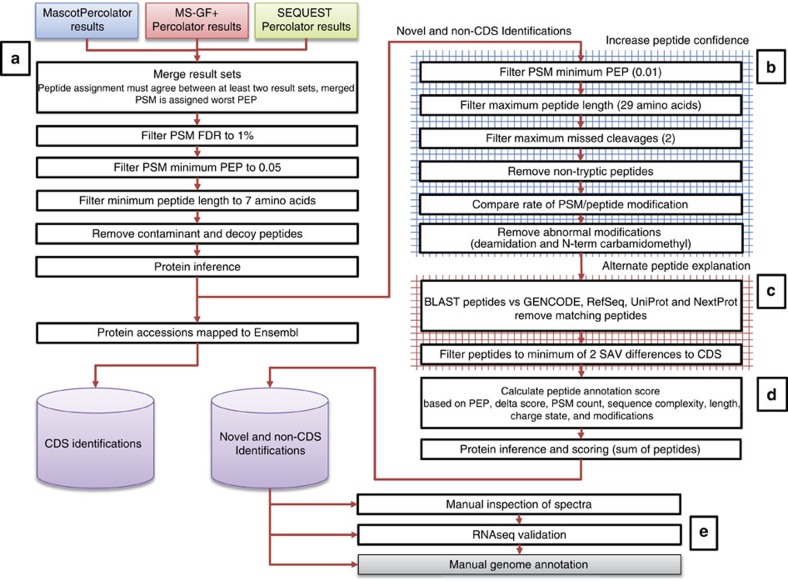
Figure 4. High-stringency peptide filtering regime.
This workflow lays out the extensive filtering applied to search results before being passed on for genome annotation. (a) Results for multiple search engines are merged requiring PSMs to match between the different engines. The worst posterior error probability (PEP) between the matching search engines is carried forward. PSMs are then filtered by false discovery rate (FDR), PEP and peptide length. Contaminants are removed prior to protein inference. Peptides with CDS matches are mapped to Ensembl and stored while novel and non-CDS only peptides are further filtered. (b) Non-CDS peptides are first filtered to increase confidence in identification. (c) These peptides are then examined for alternative explanations or existing annotation. (d) The remaining non-CDS peptides are assigned a priority annotation score and inferred into novel or non-CDS genes. (e) The final set of ranked proteins undergoes manual inspection of spectra, validation against RNAseq data sets and is passed to manual annotators for review.