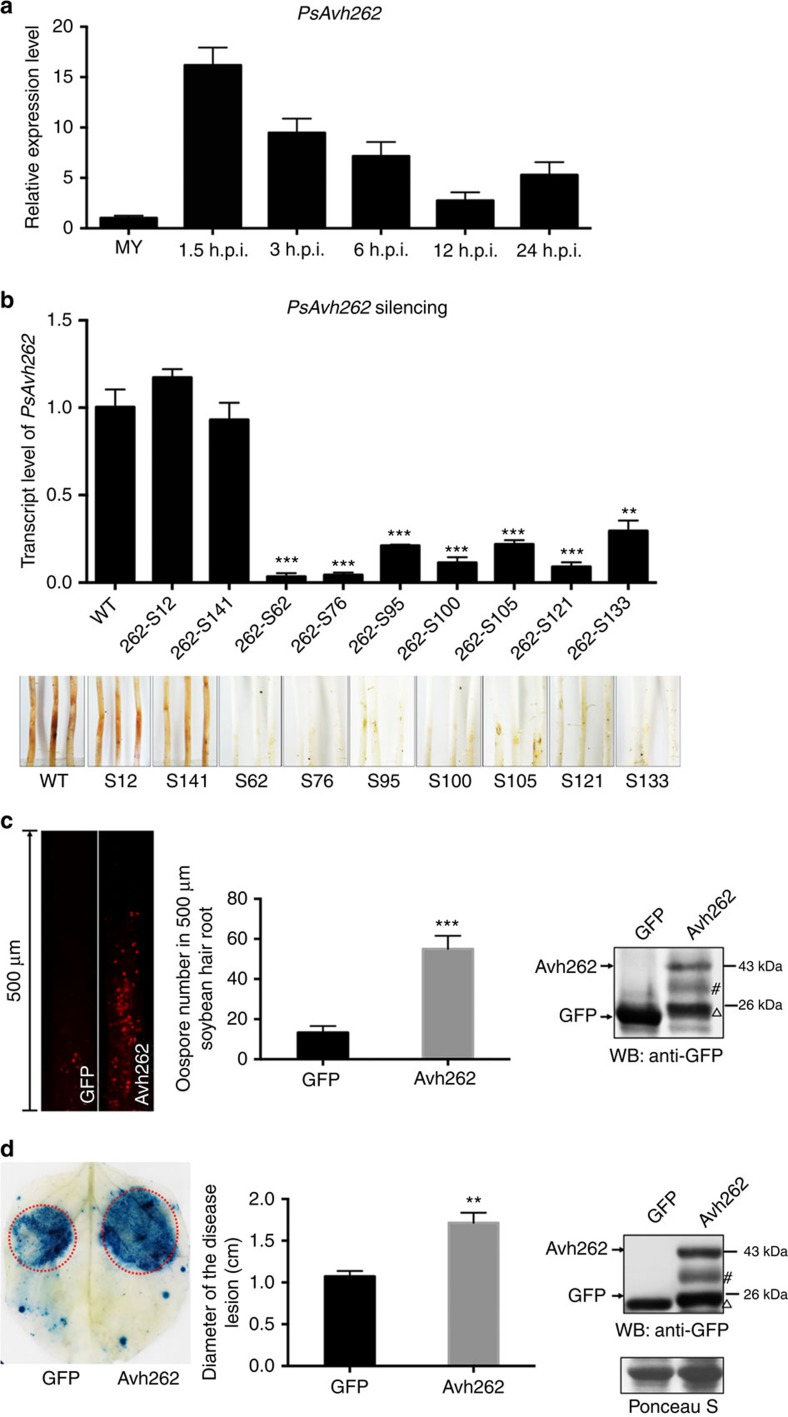
Figure 1. PsAvh262 is an essential virulence factor of Phytophthora sojae.
(a) Expression profile of PsAvh262 during P. sojae strain P6497 infection of soybean hypocotyls. The susceptible soybean cultivar Williams was used as the host. Total RNA was extracted from mycelia (MY) or infected soybean leaves at 1.5, 3, 6, 12 and 24 h post inoculation (h.p.i.). Transcript levels of PsAvh262 were determined by qRT–PCR. The P. sojae actin gene (VMD GeneID: 108986) was used as the pathogen internal control gene, (b) Silencing of PsAvh262 in P. sojae greatly impaired the virulence in soybean hypocotyls. Relative transcript levels of PsAvh262 (upper panel) in the P. sojae transformants were determined by qRT–PCR. Disease symptoms (lower panel) in etiolated hypocotyls were observed. Pictures were taken at 7 days post inoculation (d.p.i.). S12 and S141 were non-silenced transformants carrying the same silencing construct. (c) Expression of PsAvh262 in soybean hairy roots enhanced P. sojae infection. Hairy roots expressing GFP-PsAvh262 or GFP were inoculated with mycelia plugs of RFP-labelled wild-type P. sojae strain P6497 (P6497-RFP). Oospore production in the infected hair roots was observed under a confocal microscope (left panel), and lesion length was determined (middle panel) at 48 h.p.i. Expression of GFP or GFP-PsAvh262 was confirmed by western blotting using an anti-GFP antibody (right panel). (d) Expression of PsAvh262 in N. benthamiana enhanced infection of Phytophthora capsici. Leaf regions transiently expressing PsAvh262 or GFP, were inoculated with mycelia plugs of P. capsici. Infected leaves were stained with Trypan blue at 36 h.p.i. to visualize disease lesions (left panel) and the sizes of the lesions were determined (middle panel). Expression of GFP or PsAvh262-GFP was confirmed by western blotting using an anti-GFP antibody (right panel). Δ, non-specific band when using anti-GFP, which are common contaminants of western blots present in many published articles detecting GFP-fused proteins expressed in the plant cells13,64; #, PsAvh262 derived band; this may due to some unknown modification or degradation. Error bars represent the mean±s.d.(n=3) and asterisks (**or ***) denote significant differences (P<0.01 or P<0.001, assessed with one-way ANOVA). All experiments were repeated three times with similar results.