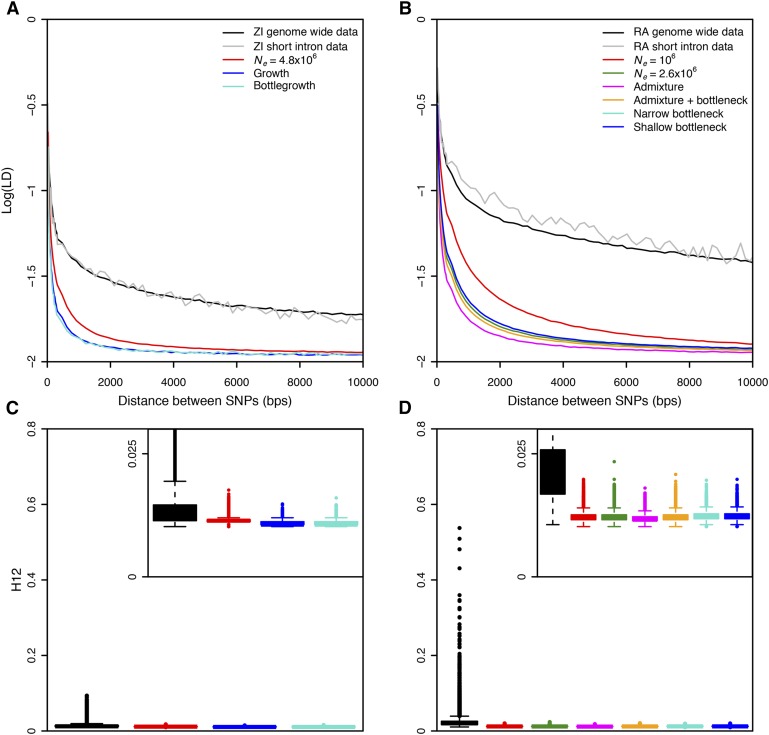
Figure 1.
Elevated long-range LD and haplotype homozygosity in Zambia and Raleigh populations. LD in (A) Zambia (ZI) and (B) Raleigh (RA) is elevated as compared to any neutral demographic model, especially for long distances. H12 values measured in (C) 801 SNP windows downsampled to 401 SNPs in Zambian data and (D) 401 SNP windows in Raleigh data were higher than expectations under any neutral demographic model tested. Pairwise LD and H12 were calculated in Zambian and Raleigh data along the regions of the autosomal arms of the D. melanogaster genome in samples of 100. Since low recombination can result in elevated LD, we excluded regions of the genome with recombination rates <1 cM/Mb (Comeron et al. 2012) and performed all neutral simulations with this recombination rate. This recombination rate should be conservative for our purposes as this was the lowest recombination rate in our data. In (A) and (B), pairwise LD was averaged over 107 simulations in each neutral demographic scenario. In (C) and (D), H12 values measured in ∼1.5 × 105 simulations under each neutral demographic model, which represents >10 times the number of analysis-observed windows in the data, are plotted.