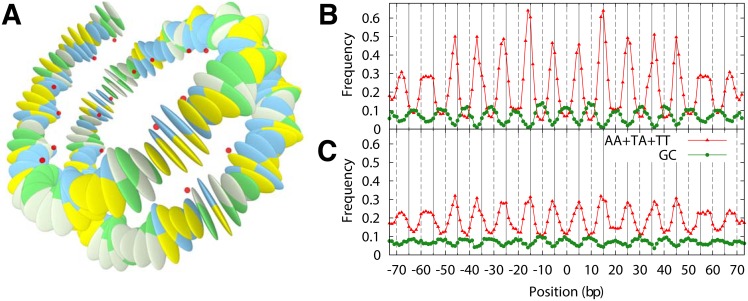
Fig 1. Nucleosomal DNA model with bp step dependent mechanical properties.
(A) The rigid base-pair model is forced, using 28 constraints (indicated by red spheres), into a lefthanded superhelical path that mimics the DNA conformation in the nucleosome crystal structure [4]. (B) Fraction of dinucleotides GC and AA/TT/TA at each position along the nucleosome model found in 10 million high affinity sequences produced by MMC at 100 K. The solid and dashed lines indicate minor and major groove bending sites; the nucleosome dyad is at 0 bp. The model recovers the basic nucleosome positioning rules [1, 3]. (C) Same as (B), but on top of 1200 coding sequences (produced by sMMC). The same periodic signals are found albeit with a smaller amplitude.