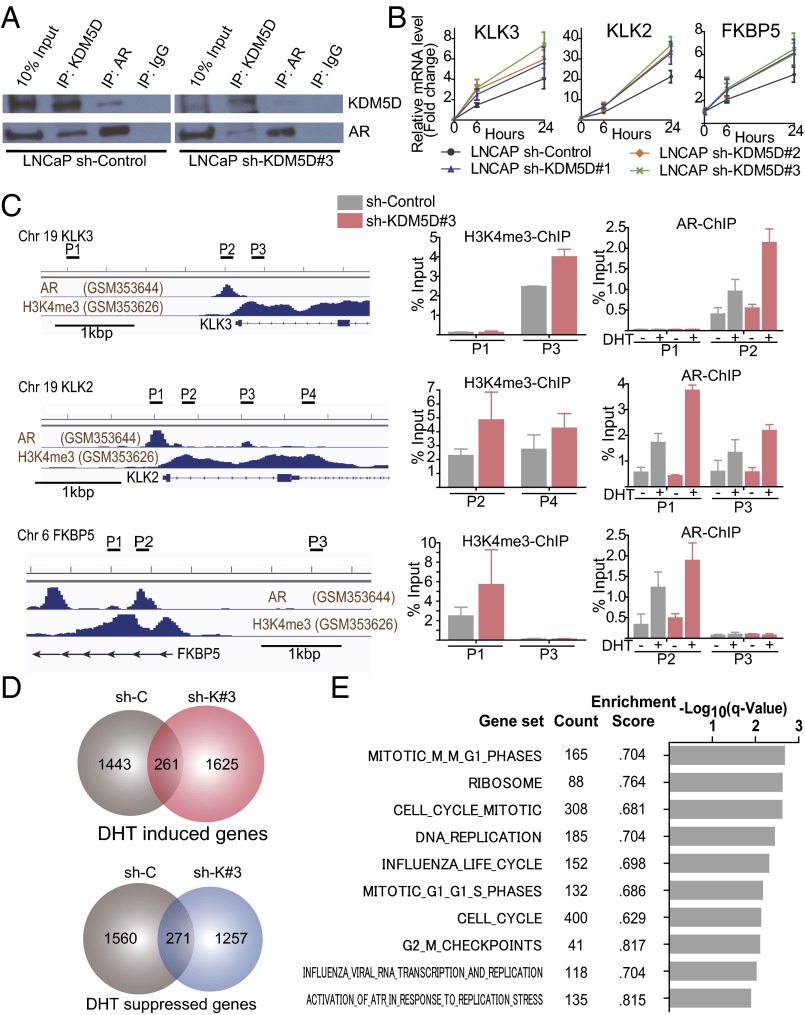
Fig. 4.
KDM5D directly interacts with AR and regulates its transcriptional activity. (A) Nuclear fractions in LNCaP sh-control and sh-KDM5D#3 cells were immunoprecipitated with antibodies specific to IgG, AR, and KDM5D followed by immunoblotting with the indicated antibodies. (B) LNCaP cells with and without shRNA to KDM5D were starved in 10% CSS media for 48 h followed by 10 nM DHT induction. RNA expression levels of AR-regulated genes were examined at 6 and 24 h after DHT by QT-PCR and normalized to GAPDH. Data are expressed as relative mean fold change compared with the expression level without DHT stimulation (mean ± SEM). (C, Left) ChIP-seq datasets of AR and H3K4me3 (27). The chromatin status in the vicinity of the transcription start site and primers designed for KLK3, KLK2, and FKBP5 are shown. (C, Right) Quantitative PCR of ChIP of H3K4me3 and AR was performed in LNCaP sh-control and sh-KDM5D#3 cells. For ChIP of H3K4me3, cells were cultured in 10% FBS for 3 d. For ChIP of AR, cells were starved for 48 h, and then treated with and without DHT (10 nM) for 16 h. Data are shown as mean ± SD. (D) Venn diagrams showing DHT-induced and -suppressed genes (gene expression fold change >2) at 24 h in LNCaP sh-control and sh-KDM5D#3. (E) Gene set enrichment analysis illustrating the top 10 enrichment gene sets up-regulated by knockdown of KDM5D in DHT (10 nM)-supplemented medium according to −log10 FDR-adjusted P value. None of the down-regulated gene sets were enriched with FDR < 0.25.