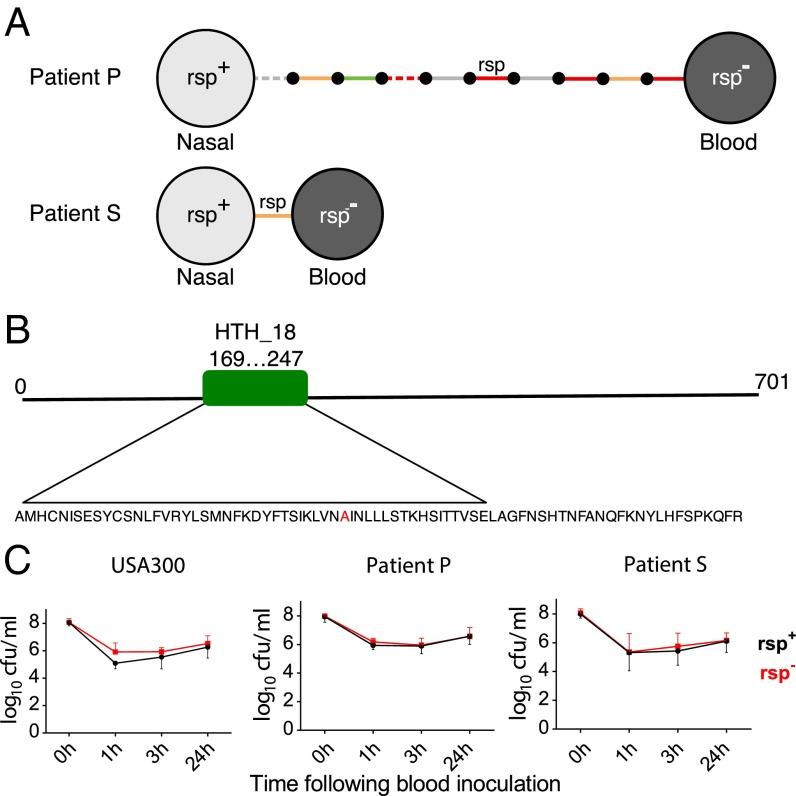
Fig. 4.
Low hemolytic rsp mutants are recovered from patients and occur naturally. (A) S. aureus from bloodstream infections carry mutations in rsp. Two bloodstream isolates were obtained from patients P and S and compared with their respective carried strains. Isolates are represented by light (rsp+) and dark (rsp−) gray circles. Intergenic (gray), synonymous (green), nonsynonymous (orange), and nonsense (red) SNPs and indels are represented by solid and dashed lines, respectively. Small black circles represent hypothetical intermediate genotypes. The ordering of mutations along the branch in patient P is arbitrary. Remarkably, only a single mutation (A204P) separated the bacteremic from a carriage isolate in patient S. (B) Rsp is highly conserved in S. aureus and contains a helix-turn-helix domain (amino acids 169–247). The observed substitution in the patient S rsp− isolate (indicated in red) occurs in the center of this domain, substituting an alanine with a proline and thereby predicted to disrupt the 3D structure of the DNA binding region. (C) Bacterial survival with the same strains used in Fig. 3, as well as with pairs of clinical isolates (A), was assessed after inoculation into human blood from three healthy donors. Bacterial survival was measured at three different time points (1, 3, and 24 h). There was no significant difference in blood survival observed between the rsp mutant (black lines) and wild-type bacteria (red lines). Statistical significance was determined by general linear modeling, modeling counts at each time point as a function of rsp genotype, and genetic background of the organism.