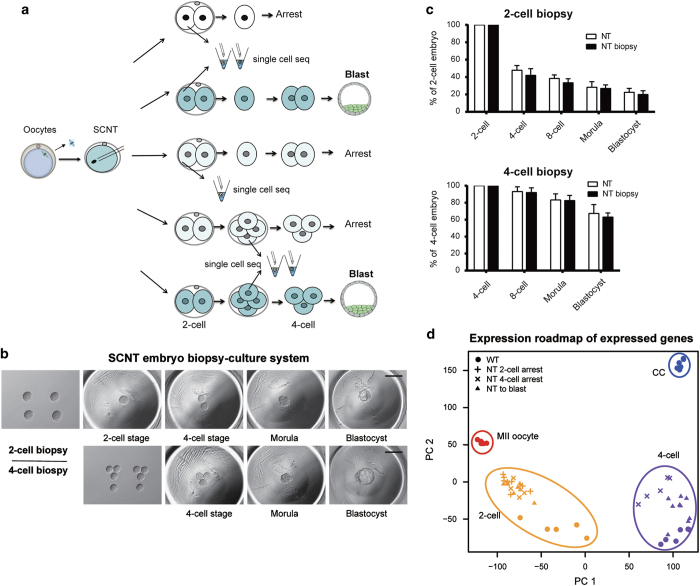
Figure 1.
Embryo biopsy enables single-cell sequencing of the SCNT embryos with distinct developmental fates. (a) Schematics of blastomere biopsy and single-cell sequencing analysis for cloned embryos with different developmental fates. One blastomere of a two- or four-cell-stage cloned embryo was picked for single-cell sequencing, and the rest were cultured to trace developmental potency. (b) Blastomere biopsy of cleavage-stage cloned embryos and culture system for developmental potency observation. Two-cell-stage (upper row) or four-cell-stage (lower row) cloned embryos were separated by gentle pipetting. After biopsy, one or three blastomeres were placed into a depression in the aggregation plate. The remaining blastomeres could arrest or develop into blastocysts. (c) Preimplantation developmental potency of cloned embryos was unaffected by blastomere biopsy. The blastocyst rates of two-cell-stage (upper panel) and four-cell-stage (lower panel) embryos after blastomere biopsy were calculated and compared with untreated SCNT embryos. The rate was calculated based on the number of blastocysts at 4.5 days post activation. The data are represented as the mean±s.d. (n>6). (d) PCA showing the genome-wide expression road map in WT and SCNT embryos with distinct cell fate. Color represents different developmental stages and shape represents different types of samples. Genes with averaged FPKM⩾1 across all samples were used in the analysis.