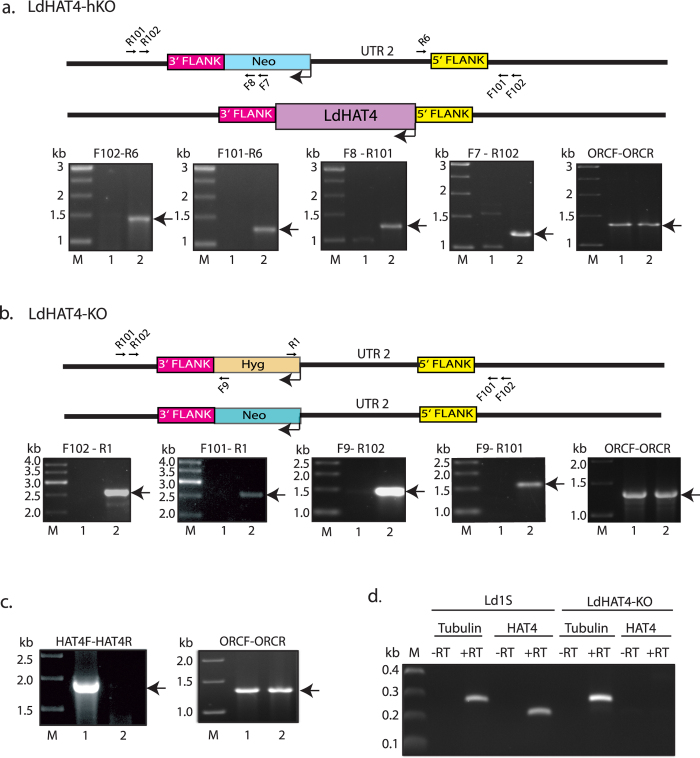
Figure 1. Creation of HAT4 knockout.
(a) Screening of heterozygous knockout line (LdHAT4-hKO) by PCRs. Primers were designed using sequences of the neor cassette, UTR2, and HAT4 flanks in the Leishmania genome. Primer positions are marked in the line diagram. Primer pair used in each PCR is indicated above the agarose gel image. OrcF - OrcR PCR: positive control for templates. Lanes 1- Ld1S genomic DNA, lanes 2- LdHAT4-hKO genomic DNA, lanes M- DNA ladder. (b) Screening of knockout line (LdHAT4-KO) by PCRs. Primers were designed using sequences of the hygr cassette and HAT4 flanks in the Leishmania genome; positions of primers are marked in the line diagram; primer pair used in each PCR is indicated above gel image. Lanes 1- Ld1S genomic DNA, lanes 2- LdHAT4-KO genomic DNA, lanes M- DNA ladder. (c) Confirmation of HAT4 deletion in LdHAT4-KO line. PCRs carried out using HAT4 primers, with Ld1S genomic DNA (lane 1) and LdHAT4-KO genomic DNA (lane 2) as templates. (d) Analysis of HAT4 transcription in Ld1S and LdHAT4-KO cells. –RT: non-reverse transcribed RNA as template (control for checking genomic DNA contamination). +RT: reverse transcribed RNA as template. Positive control: reactions with tubulin primers.