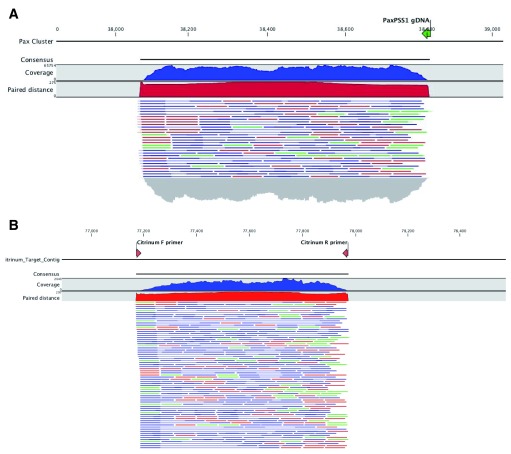
Figure 4. PCR of genes encoding Paxilline and Citrinin demonstrates amplification of the expected size.
Citrinum primers we designed from Genbank accession number LKUP01000753. Paxilline primers were used as described in Saikia et al. PCR products were made into shotgun libraries with Nextera and sequenced on an Illumina MiSeq with 2×75bp reads to over 10,000X coverage. Reads were mapped with CLCbio 4 to NCBI accession number HM171111.1 ( A) and LKUP01000000 respectively ( B). Paired reads are displayed as blue lines, green and red lines are unpaired reads. Read coverage over the amplicons are depicted in a blue histogram over the cluster while paired end read distance is measured in a red histogram over the region. Off target read mapping is limited. P. paxilli mappings are displayed on top ( A) and P. citrinum mappings are displayed on bottom ( B). Alignment of PCR primers to P. paxilli reference shows a 5 prime mismatch that is a result of the primers being designed to target spliced RNA according to Saikia et al.