Fig. 1.
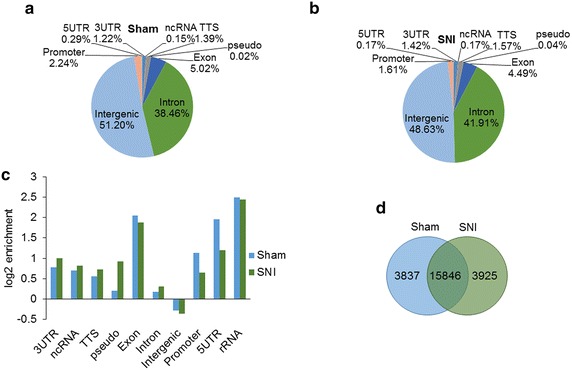
Genome-wide MeCP2-DNA binding profile in the DRG from SNI and sham control mice. a–b Genomic distribution of peaks from ChIP-seq in the sham and SNI samples. c Enrichment of MeCP2 binding to genomic regions in the SNI and sham models. Enrichment in SNI and sham, and p values are shown in Table 1. d Common and unique broad peaks identified in regions encoding mRNAs in SNI and sham. ChIP-seq data were generated from two IP samples each, for SNI and sham. Each sample contained L4, L5 and L6 ipsilateral DRG obtained from ten mice each, 4 weeks after surgery. 3UTR 3′ untranslated region, 5UTR 5′ untranslated region, ncRNA noncoding RNA, Pseudo pseudo genes, rRNA ribosomal RNA, TTS transcription termination sites
