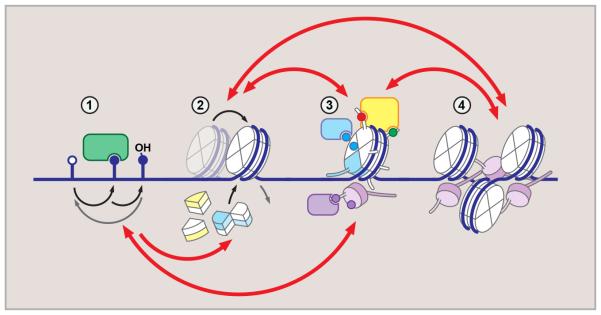
Figure 1. Hierarchical organization of epigenetic regulation.
Dynamic and reversible epigenetic processes generate diverse regulatory environment. DNA methylation (1) (mC, methylcytosine – closed circle) may result in eviction of DNA-binding proteins, or recruitment of methyl-binding factors. mC oxidation (shown is hydroxymethylcytosine, hmC) generates additional diversity. Core and linker histone exchange, including variant histone incorporation (2), regulate local DNA accessibility, and, together with histone modifications (3), introduce local variations to chromatin structure. These are interpreted by the “reader” machinery (3) and drive higher-order chromatin organization and nuclear topology (4). Red arrows above and below indicate crosstalk between regulatory layers.