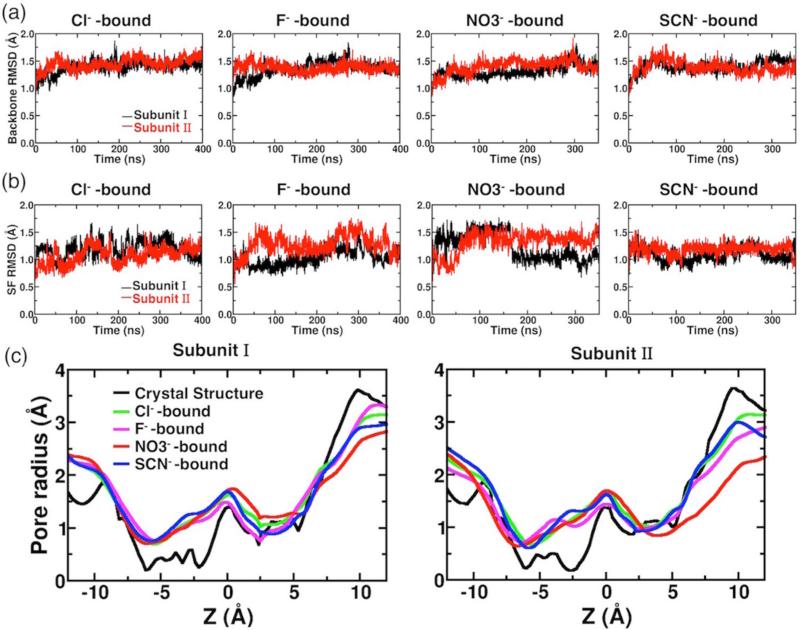
Figure 2.
Protein structural dynamics in the presence of different anions. (a) Backbone RMSDs of individual subunit shown as a function of simulation time for anion-bound simulations. (b) The RMSDs of all heavy (non-hydrogen) atoms of the selectivity filter (SF) in the presence of different anions. (c) Comparison of pore radius profiles of the crystal structure (black) and those of MD simulations (averaged over the whole trajectory) with different anions bound at Scen (colored traces). The center of the bound anion at t = 0, which corresponds to the Cl− position in the crystal structure, is set as the origin. The extracellular side is toward the left (z < 0), and the intracellular side is toward the right (z > 0).