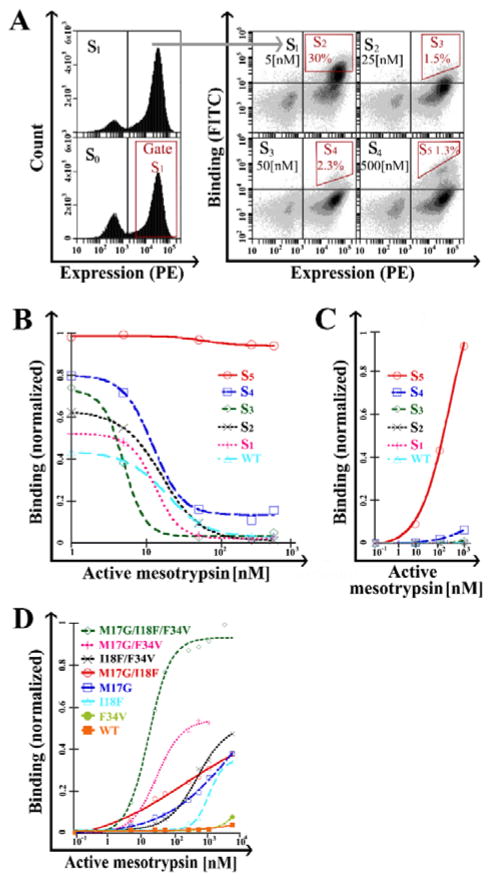
Figure 2. Identification of APPI clones with improved resistance to cleavage.
(A) Stability maturation of the APPI library. FACS of single or dual-labeled cell populations for expression (S0 and S1) or both expression and binding (S1 to S5). The expressed population of APPI variants was sorted (S0), and the expression of the library was tested after enrichment (S1). Next, each cycle of stability maturation (S2 to S5) was performed with elevated concentrations of active mesotrypsin (as noted in the upper right quadrant of each plot) and a fixed concentration of inactive mesotrypsin (2 μM). Sorting gates are marked in red. (B) ‘Triple staining’ and (C) ‘double staining’ analysis of cell populations from library maturation cycles. (D) ‘Double staining’ analysis of cells expressing M17G, I18F, F34V and combination variants. A leftward shift in the sigmoid shape indicates a higher affinity, whereas a higher binding signal under saturating conditions indicates higher proteolytic stability. In panels B–D, the Y-axis represents mean fluorescence intensity normalization of binding to expression. Data was analyzed using KaleidaGraph software, with a sigmoidal curve fit. For all panels, the surface expression of APPI was detected by using a primary antibody against the C-terminal c-Myc tag and a PE-labeled secondary antibody, while binding to APPI was detected by biotinylated mesotrypsin and FITC-labeled streptavidin.