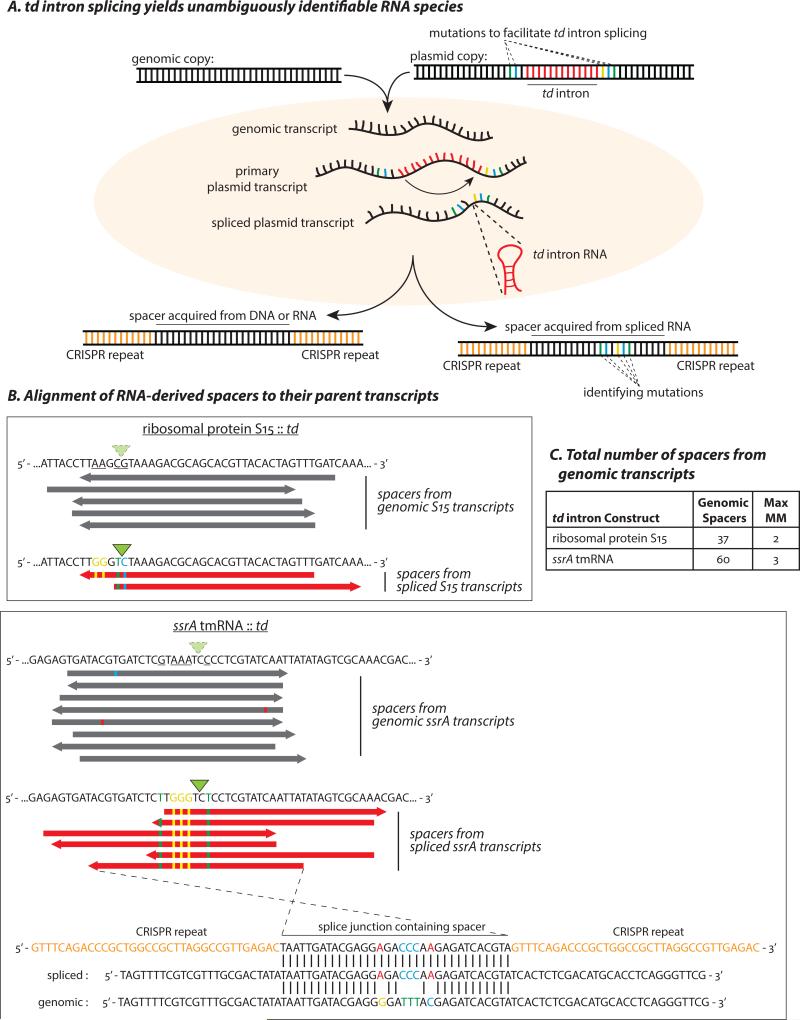
Figure 4. Spacer acquisition from RNA in the MMB-1 Type III-B system.
(A) Spacers acquired from a host genome could conceivably originate from either RNA or DNA. To test for an RNA origin, we used an engineered self-splicing transcript, which produces an RNA sequence junction that is not encoded by DNA. Bases that were mutated to provide flanking exon sequences favorable for td intron splicing are separated by the 393 bp intron in the DNA template. Following transcription and splicing, the two exons are brought together to form a novel junction containing the “identifying mutations”. Newly acquired spacers that contain this exon-junction indicate spacer acquisition from an RNA target. (B) Alignments of some of the genome-contiguous spacers (gray) and several newly acquired exon-junction spanning spacers (red) to the genomic and split-gene sequences, respectively. Bases mutated to facilitate td intron splicing are underlined in the genomic sequences. Identifying mutations are depicted as colored bases, and the splice sites are indicated by green triangles. The highlighted ssrA exon-junction spanning spacer (bottom) is antisense to the spliced tmRNA and differs from a putative DNA template by the 5 expected mutations. (C) All unique spacers spanning the td-intron splice site that did not carry the engineered mutations. The maximum number of mismatches when these spacers were mapped to the wild-type genomic locus is indicated. None of the identifying mutations were observed among these sporadic mismatches. The spacers in (B) were in addition to four spacers (1 for the S15 and 3 for the ssrA construct) that align to the unspliced exon-intron junction and could have been derived from either DNA or (nascent) RNA.