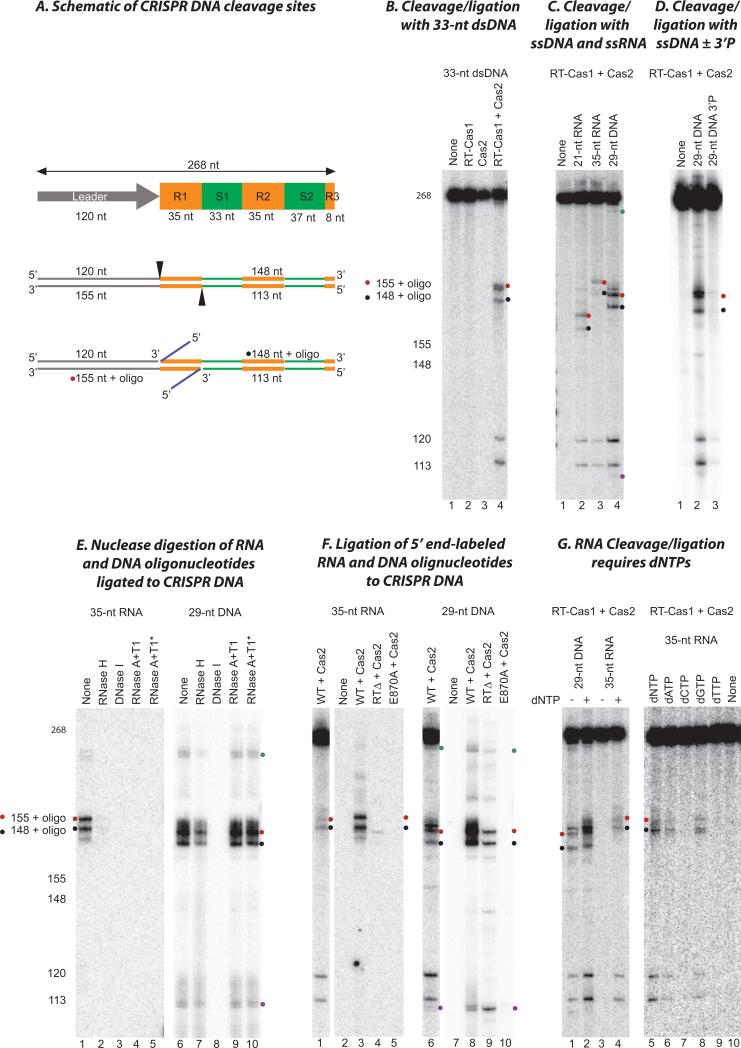
Figure 5. Site-specific CRISPR DNA cleavage/ligation by RT-Cas1/Cas2.
(A) Schematic of CRISPR DNA substrates and products of cleavage/ligation reactions. The substrate was a 268 bp DNA containing the leader (gray), the first two repeats (R1 and R2; orange) and spacers (S1 and S2; green), and part of the third repeat (orange) of the MMB-1 CRISPR03 array. Cleavages (arrowheads) occur at the boundaries of the first repeat with concomitant ligation of a DNA or RNA oligonucleotide (blue) to the 3’ fragment, yielding products of the sizes shown. (B) Internally labeled CRISPR DNA and a 33-nt dsDNA were incubated with no protein (None, lane 1), RT-Cas1 (lane 2), Cas2 (lane 3), or a 1:2 mixture of RT-Cas1 and Cas2 (lane 4). The sizes of products determined from sequencing ladders in parallel lanes are indicated (left). (C) Internally labeled CRISPR DNA was incubated with WT RT-Cas1 and Cas2 without (lane 1) or with a 21-nt RNA (lane 2), 35-nt RNA (lane 3), or 29-nt ssDNA (lane 4). (D) Internally labeled CRISPR DNA was incubated with WT RT-Cas1+Cas2 in the absence (none), or presence of a 29-nt ssDNA with either a 3’ OH (lane 2) or a 3’ phosphate (lane 3). (E) Nuclease digestion of 5’ end-labeled RNA and DNA oligonucleotides ligated to CRISPR DNA. Ligation reactions were done as in (C). After extraction with phenol-CIA and ethanol precipitation, the products were incubated with the indicated nucleases. An asterisk indicates that the sample was boiled to denature the DNA before adding the nuclease. (F) Ligation of 5’ end-labeled RNA and DNA oligonucleotides into CRISPR DNA by WT and mutant RT-Cas1 proteins. Lanes 1 and 6 show control reactions of internally labeled CRISPR with WT RT-Cas1+Cas2 and an unlabeled 35-nt ssRNA or 29-nt ssDNA oligonucleotide for comparison. Lanes 2-5 and 7-10 show reactions of unlabeled CRISPR DNA with 5’-end labeled 35-nt ssRNA and 29-nt ssDNA, respectively, and WT, E870A, and RTΔ RT-Cas1 plus Cas2. All reactions were done in the presence of dNTPs. (G) Effect of dNTPs. In the gel to the left, internally labeled CRISPR DNA was incubated with WT RT-Cas1 plus Cas2 in the presence of a 29-nt ssDNA (lanes 1 and 2) or 35-nt ssRNA (lanes 3 and 4) in the absence (lanes 1 and 3) or presence of 1 mM dNTPs (1 mM each of dATP, dCTP, dGTP, and dTTP; lanes 2 and 4). In the gel to the right, internally labeled CRISPR DNA was incubated with WT RT-Cas1+Cas2 in the presence of a 35-nt ssRNA oligonucleotide in the absence (none, lane 10) or presence of different dNTPs (1 mM) as indicated (lanes 5 to 9). Red and black dots indicate products resulting from cleavage and ligation of oligonucleotides at the junction of the leader and first repeat on the top strand and the junction of repeat 1 and spacer 1 on the bottom strand, respectively; cyan and purple dots indicate products of the size expected for cleavage and ligation of the oligonucleotide at the junctions of the second CRISPR repeat (see Fig. S10).