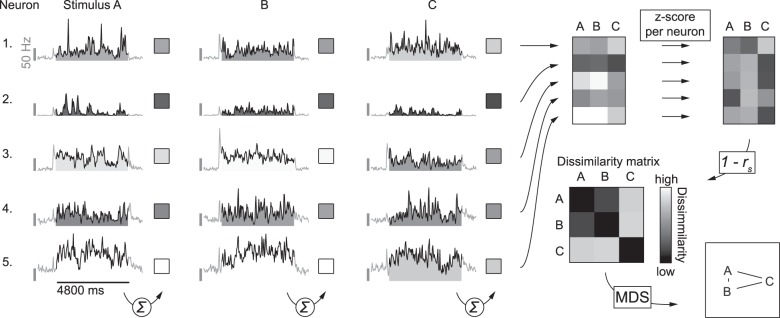
Figure 2.
Schematic illustration of how dissimilarity matrices were calculated. For the analysis of the population representation we started with the responses of SUs, averaged across repeated stimulus presentations and summed across the 201 to 5000 ms window calculated from response onset (as indicated by the summation symbol). PSTH's in this example figure illustrate responses averaged across stimulus presentations of 5 SUs to 3 movies (A–C), all arbitrarily selected for the purpose of illustrating the methods. This was done for each stimulus (e. g. A–C, in this example) to get raw response vectors. These raw responses were further standardized per neuron, by calculating the Z-scores across stimuli. Next the correlation matrix was calculated from these normalized response data by pairwise correlation of the stimulus response vectors. For the stimulus dissimilarity matrix each value in this correlation matrix was subtracted from one, resulting in values between 0 and 2, where 0 indicates the lowest dissimilarity (i.e., an identical population response pattern) and 2 indicates the highest dissimilarity (i.e., a highly different population response pattern). To visualize the stimulus space as represented by the population of neurons, we performed multidimensional scaling on the dissimilarity matrix and present the stimuli using the first 2 dimensions. Stimuli plotted closer together (A,B in this example) have a more similar population response pattern than stimuli plotted further apart (A,C, and B,C in this example).