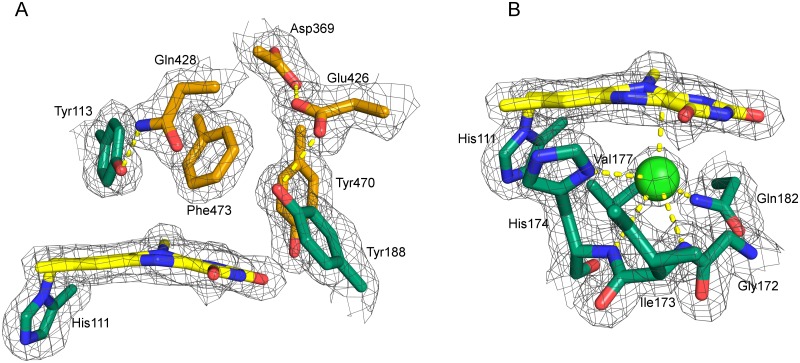
Fig 3. A: Amino acid residues present on the si-side (A) and re-side (B) of the isoalloxazine ring.
A: Residues originating from the FAD- and substrate binding domain are shown in green and orange, respectively. The 2Fo-Fc is shown at 1.5σ. The carboxylate groups of Asp369 and Glu426 are found at a distance of 2.4 Å. The proximity of these residues suggests a shared proton and a negative charge delocalized over both carboxylic acids. Glu426 interacts with Tyr188 that putatively acts as catalytic base after de-protonation by Glu426. B: Residues defining the re-side of the isoalloxazine ring. An oxygen pocket is formed by the oxygen reactivity motif (compare Fig 2B). The electron density was interpreted as a chloride ion (light green), which is complexed by the backbone amides of His174 and Ile173, as well as Nπ of His174 and the side chain amide of Gln182.