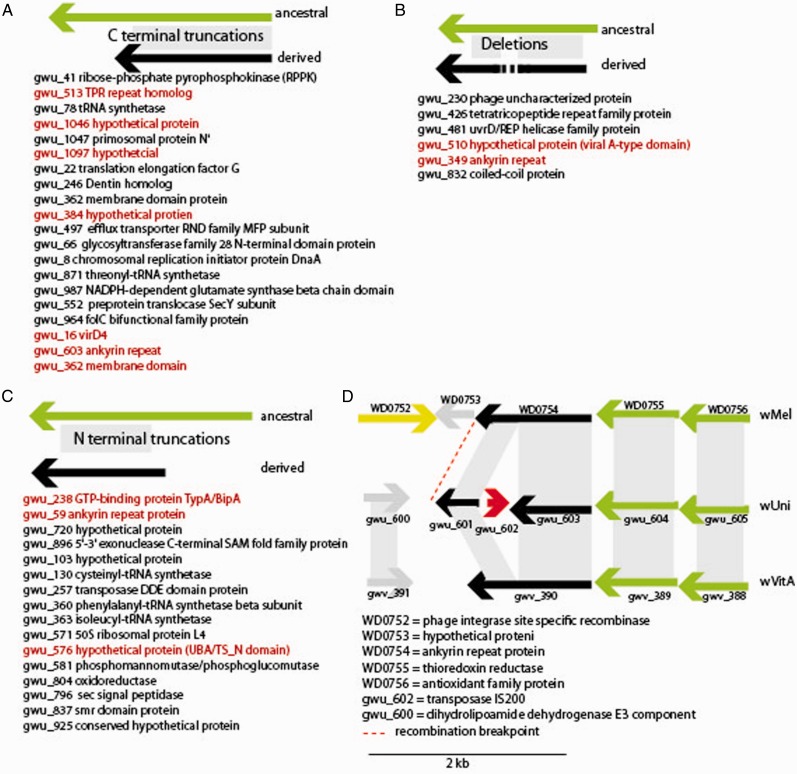
Fig. 4.—
Schematic representation of gene evolution by truncations (A and C) and deletions (B) in the wUni genome with respect to the type A Wolbachia (wVitA, wMel, and wRi), some of which result from genomic rearrangements. Red text denotes those proteins predicted to be involved in host interaction based on secretion signal, a eukaryotic domain (such as an ankyrin repeat domain), and conservation of ancestral state with wHa, wRi, and wVitA. Only three wVitA genes (gwv_46, gwv_937, gwv_909) are predicted to have undergone similar truncations and deletions relative to the outgroup type A Wolbachia. (D) Illustrative example gene truncation due to transposon insertion within an ankyrin repeat protein. The wUni gene gwu_602 encodes an IS200 element that has inserted within an ankyrin repeat domain protein found intact in both the wVitA (gwv_390) and wMel genomes (WD0754). This segment of the genome is syntenic between these three type A Wolbachia strains.